使用发散诊断有偏推断#
from collections import defaultdict
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pymc3 as pm
print(f"Running on PyMC3 v{pm.__version__}")
Running on PyMC3 v3.11.5
%config InlineBackend.figure_format = 'retina'
az.style.use("arviz-darkgrid")
SEED = [20100420, 20134234]
此笔记本是 PyMC3 对 Michael Betancourt 在 mc-stan 上的帖子的移植。有关底层机制的详细解释,请查看原始帖子 使用发散诊断有偏推断 和 Betancourt 的优秀论文 哈密顿蒙特卡罗的概念性介绍。
贝叶斯统计完全是关于构建模型和估计模型中的参数。但是,我们概率模型的朴素或直接参数化有时可能无效,您可以查看 Thomas Wiecki 的博客文章 为什么分层模型很棒、棘手且是贝叶斯模型,了解 PyMC3 中的相同问题。次优的参数化通常会导致缓慢的抽样,更成问题的是,会导致有偏差的 MCMC 估计量。
更正式地说,正如原始帖子 使用发散诊断有偏推断 中解释的那样
马尔可夫链蒙特卡罗 (MCMC) 近似于关于给定目标分布的期望,
使用马尔可夫链的状态,\({q{0}, \ldots, q_{N} }\),
然而,这些估计量仅在链增长到无限长时才保证是准确的,
为了在应用分析中发挥作用,我们需要 MCMC 估计量足够快地收敛到真实期望值,以便在我们耗尽有限的计算资源之前,它们是相当准确的。这种快速收敛需要强大的遍历性条件成立,特别是马尔可夫转移和目标分布之间的几何遍历性。几何遍历性通常是 MCMC 估计量遵循中心极限定理的必要条件,这不仅确保即使在有限次迭代后它们也是无偏的,而且还确保我们可以使用 MCMC 标准误差凭经验量化它们的精度。
不幸的是,对于任何非平凡的问题,证明几何遍历性都是不可行的。相反,我们必须依赖经验诊断来识别对几何遍历性的阻碍,从而识别表现良好的 MCMC 估计量。对于一般的马尔可夫转移和目标分布,最著名的诊断是在从参数空间中的扩散点初始化的马尔可夫链集合上的分裂 \(\hat{R}\) 统计量;为了做得更好,我们需要利用给定转移或目标分布的特定结构。
例如,哈密顿蒙特卡罗在这方面尤其强大,因为它未能关于任何目标分布在几何上是遍历的,这表现在已发展成敏感诊断的不同行为中。其中一种行为是发散的出现,这表明哈密顿马尔可夫链遇到了目标分布中曲率高的区域,它无法充分探索这些区域。
在本笔记本中,我们的目标是识别 PyMC3 中的发散和潜在的病理。
八所学校模型#
八所学校数据集(Rubin 1981)的分层模型,如 Stan 中所示
其中 \(n \in \{1, \ldots, 8 \}\) 且 \(\{ y_{n}, \sigma_{n} \}\) 作为数据给出。
推断分层超参数 \(\mu\) 和 \(\sigma\),以及组级参数 \(\theta_{1}, \ldots, \theta_{8}\),允许模型跨组汇集数据并减少其后验方差。不幸的是,直接中心化参数化还将后验分布压缩成特别具有挑战性的几何形状,从而阻碍了几何遍历性,并因此使 MCMC 估计产生偏差。
中心化八所学校实施#
Stan 模型
data {
int<lower=0> J;
real y[J];
real<lower=0> sigma[J];
}
parameters {
real mu;
real<lower=0> tau;
real theta[J];
}
model {
mu ~ normal(0, 5);
tau ~ cauchy(0, 5);
theta ~ normal(mu, tau);
y ~ normal(theta, sigma);
}
同样,我们可以很容易地在 PyMC3 中实现它
with pm.Model() as Centered_eight:
mu = pm.Normal("mu", mu=0, sigma=5)
tau = pm.HalfCauchy("tau", beta=5)
theta = pm.Normal("theta", mu=mu, sigma=tau, shape=J)
obs = pm.Normal("obs", mu=theta, sigma=sigma, observed=y)
不幸的是,模型的这种直接实现表现出病态的几何形状,从而阻碍了几何遍历性。更令人担忧的是,由此产生的偏差是微妙的,并且在单独检查马尔可夫链时可能并不明显。为了理解这种偏差,让我们首先考虑一个短马尔可夫链,当计算上的便利性是一个驱动因素时通常使用,然后再考虑一个更长的马尔可夫链。
危险的短马尔可夫链#
with Centered_eight:
short_trace = pm.sample(600, chains=2, random_seed=SEED)
/Users/reshamashaikh/miniforge3/envs/pymc-ex/lib/python3.10/site-packages/deprecat/classic.py:215: FutureWarning: In v4.0, pm.sample will return an `arviz.InferenceData` object instead of a `MultiTrace` by default. You can pass return_inferencedata=True or return_inferencedata=False to be safe and silence this warning.
return wrapped_(*args_, **kwargs_)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [theta, tau, mu]
Sampling 2 chains for 1_000 tune and 600 draw iterations (2_000 + 1_200 draws total) took 16 seconds.
There were 52 divergences after tuning. Increase `target_accept` or reparameterize.
The acceptance probability does not match the target. It is 0.4129320535021329, but should be close to 0.8. Try to increase the number of tuning steps.
There were 10 divergences after tuning. Increase `target_accept` or reparameterize.
The acceptance probability does not match the target. It is 0.6090970402923143, but should be close to 0.8. Try to increase the number of tuning steps.
The rhat statistic is larger than 1.4 for some parameters. The sampler did not converge.
The estimated number of effective samples is smaller than 200 for some parameters.
在 原始帖子 中,应用了 1200 个样本的单链。但是,由于 PyMC3 中未实现拆分 \(\hat{R}\),因此我们改为拟合 2 个链,每个链包含 600 个样本。
Gelman-Rubin 诊断 \(\hat{R}\) 未指示任何问题(值都接近 1)。您可以尝试使用不同的种子重新运行模型,看看这是否仍然成立。
az.summary(short_trace).round(2)
Got error No model on context stack. trying to find log_likelihood in translation.
/Users/reshamashaikh/miniforge3/envs/pymc-ex/lib/python3.10/site-packages/arviz/data/io_pymc3_3x.py:98: FutureWarning: Using `from_pymc3` without the model will be deprecated in a future release. Not using the model will return less accurate and less useful results. Make sure you use the model argument or call from_pymc3 within a model context.
warnings.warn(
| 均值 | 标准差 | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| mu | 3.76 | 2.84 | -2.00 | 9.43 | 0.20 | 0.15 | 182.0 | 288.0 | 1.20 |
| theta[0] | 5.29 | 4.88 | -4.38 | 14.48 | 0.30 | 0.32 | 220.0 | 445.0 | 1.28 |
| theta[1] | 4.33 | 4.28 | -3.78 | 13.19 | 0.25 | 0.27 | 257.0 | 275.0 | 1.40 |
| theta[2] | 3.20 | 4.64 | -6.18 | 12.93 | 0.26 | 0.25 | 254.0 | 437.0 | 1.10 |
| theta[3] | 4.04 | 4.23 | -4.63 | 12.05 | 0.22 | 0.20 | 247.0 | 402.0 | 1.12 |
| theta[4] | 3.11 | 4.10 | -5.22 | 11.27 | 0.21 | 0.17 | 292.0 | 290.0 | 1.18 |
| theta[5] | 3.44 | 4.47 | -7.27 | 11.66 | 0.24 | 0.38 | 289.0 | 327.0 | 1.38 |
| theta[6] | 5.36 | 4.35 | -2.80 | 14.17 | 0.33 | 0.33 | 175.0 | 395.0 | 1.25 |
| theta[7] | 4.17 | 4.55 | -5.80 | 12.50 | 0.23 | 0.19 | 328.0 | 455.0 | 1.47 |
| tau | 3.26 | 2.78 | 0.62 | 8.13 | 1.01 | 0.74 | 4.0 | 6.0 | 1.58 |
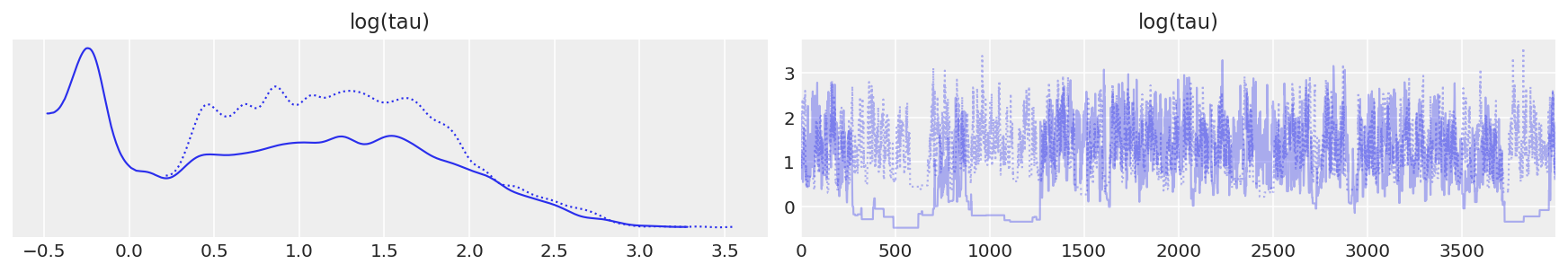
此外,迹图看起来都不错。例如,让我们考虑分层标准差 \(\tau\),或者更具体地说,它的对数 \(log(\tau)\)。由于 \(\tau\) 被约束为正数,因此它的对数将使我们能够更好地解决小值的行为。实际上,链似乎在合理地探索小值和大值。
# plot the trace of log(tau)
ax = az.plot_trace(
{"log(tau)": short_trace.get_values(varname="tau_log__", combine=False)}, legend=True
)
ax[0, 1].set_xlabel("Draw")
ax[0, 1].set_ylabel("log(tau)")
ax[0, 1].set_title("")
ax[0, 0].set_xlabel("log(tau)")
ax[0, 0].set_title("Probability density function of log(tau)");
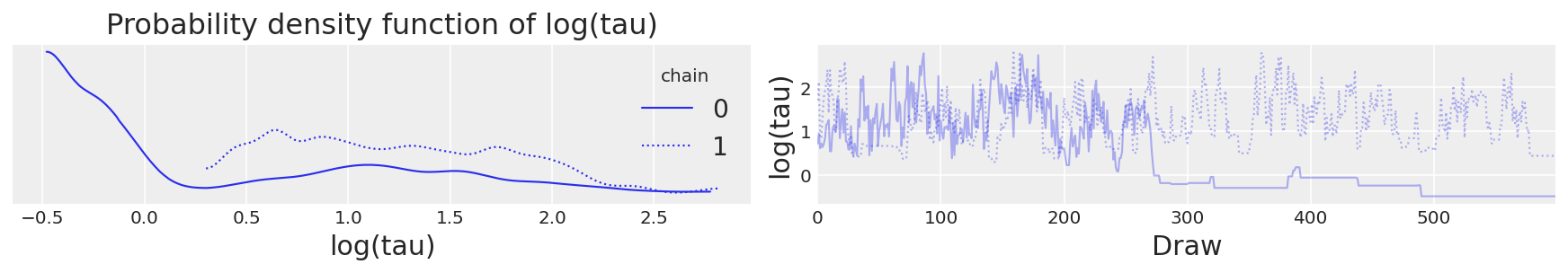
log(tau) 的迹图#
不幸的是,\(log(\tau)\) 均值的最终估计值与真实值(此处以灰色显示)存在很大的偏差。
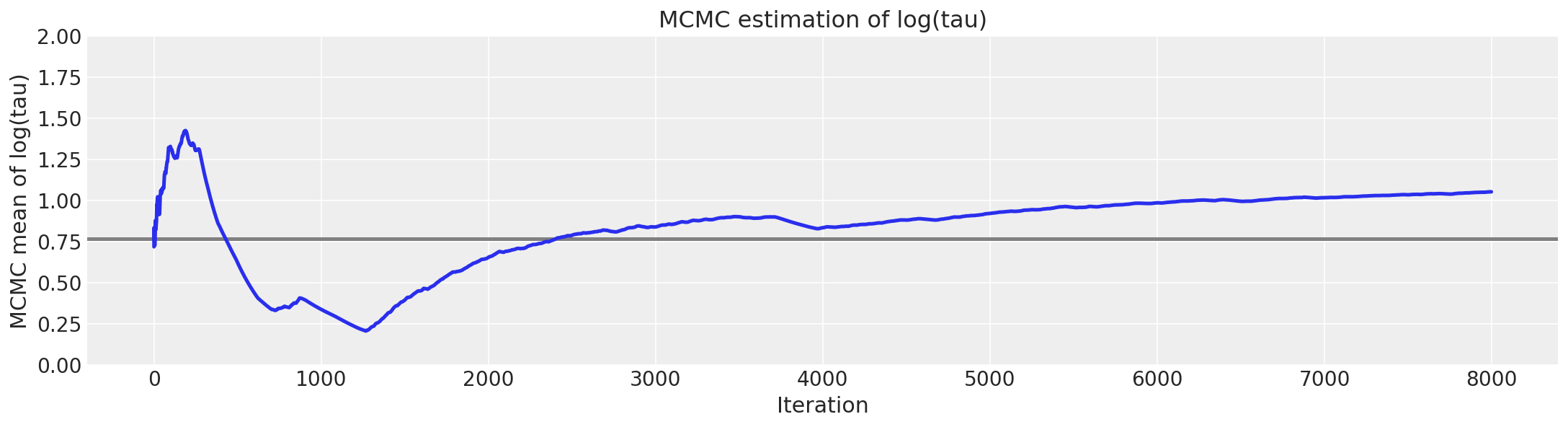
# plot the estimate for the mean of log(τ) cumulating mean
logtau = np.log(short_trace["tau"])
mlogtau = [np.mean(logtau[:i]) for i in np.arange(1, len(logtau))]
plt.figure(figsize=(15, 4))
plt.axhline(0.7657852, lw=2.5, color="gray")
plt.plot(mlogtau, lw=2.5)
plt.ylim(0, 2)
plt.xlabel("Iteration")
plt.ylabel("MCMC mean of log(tau)")
plt.title("MCMC estimation of log(tau)");
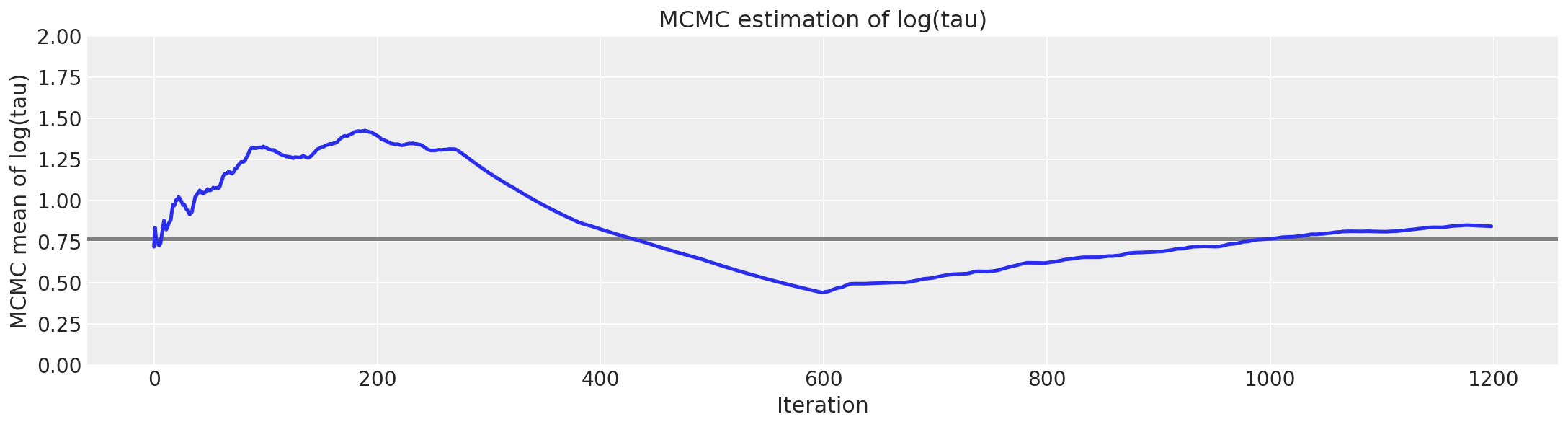
但是,哈密顿蒙特卡罗并非对这些问题一无所知,因为我们唯一的马尔可夫链中约有 3% 的迭代以发散结束。
# display the total number and percentage of divergent
divergent = short_trace["diverging"]
print("Number of Divergent %d" % divergent.nonzero()[0].size)
divperc = divergent.nonzero()[0].size / len(short_trace) * 100
print("Percentage of Divergent %.1f" % divperc)
Number of Divergent 62
Percentage of Divergent 10.3
即使使用单个短链,这些发散也能够识别偏差,并建议对任何由此产生的 MCMC 估计量持怀疑态度。
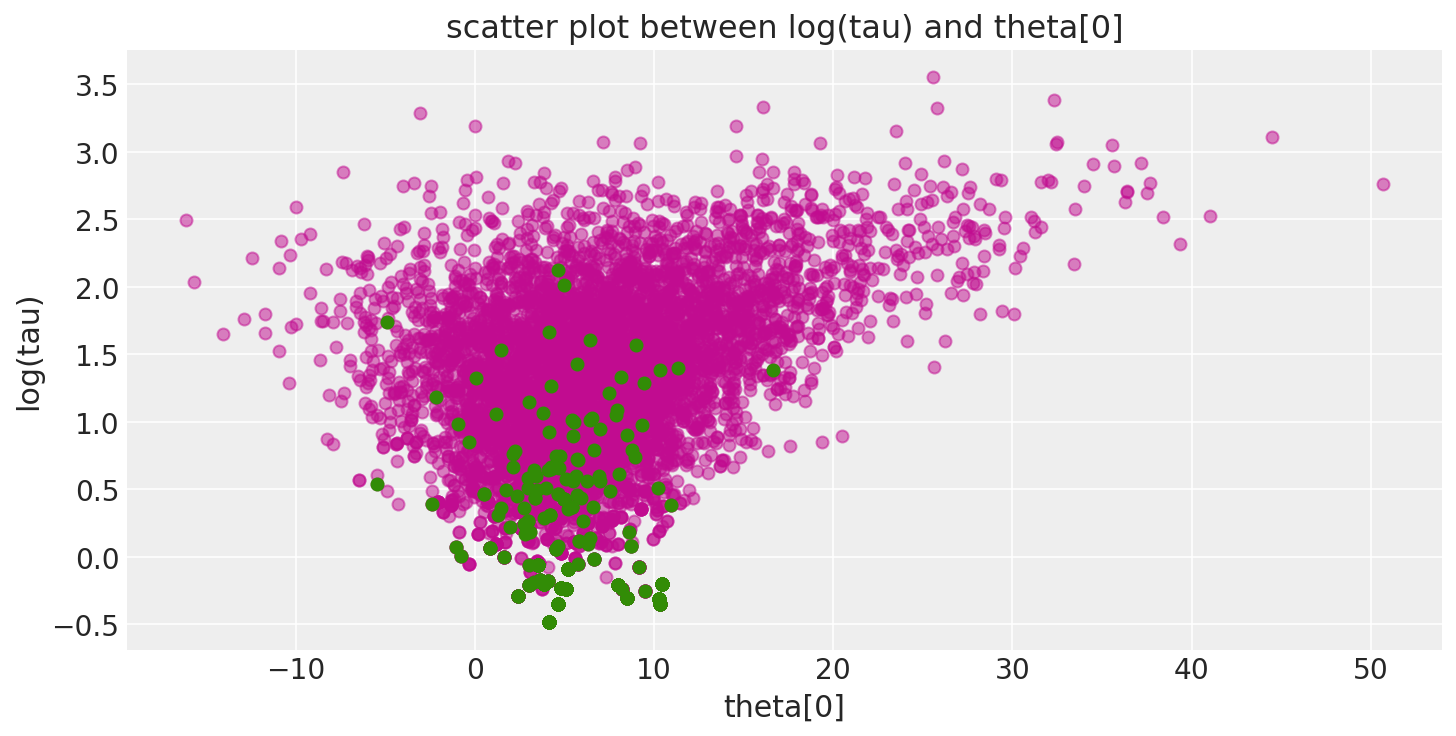
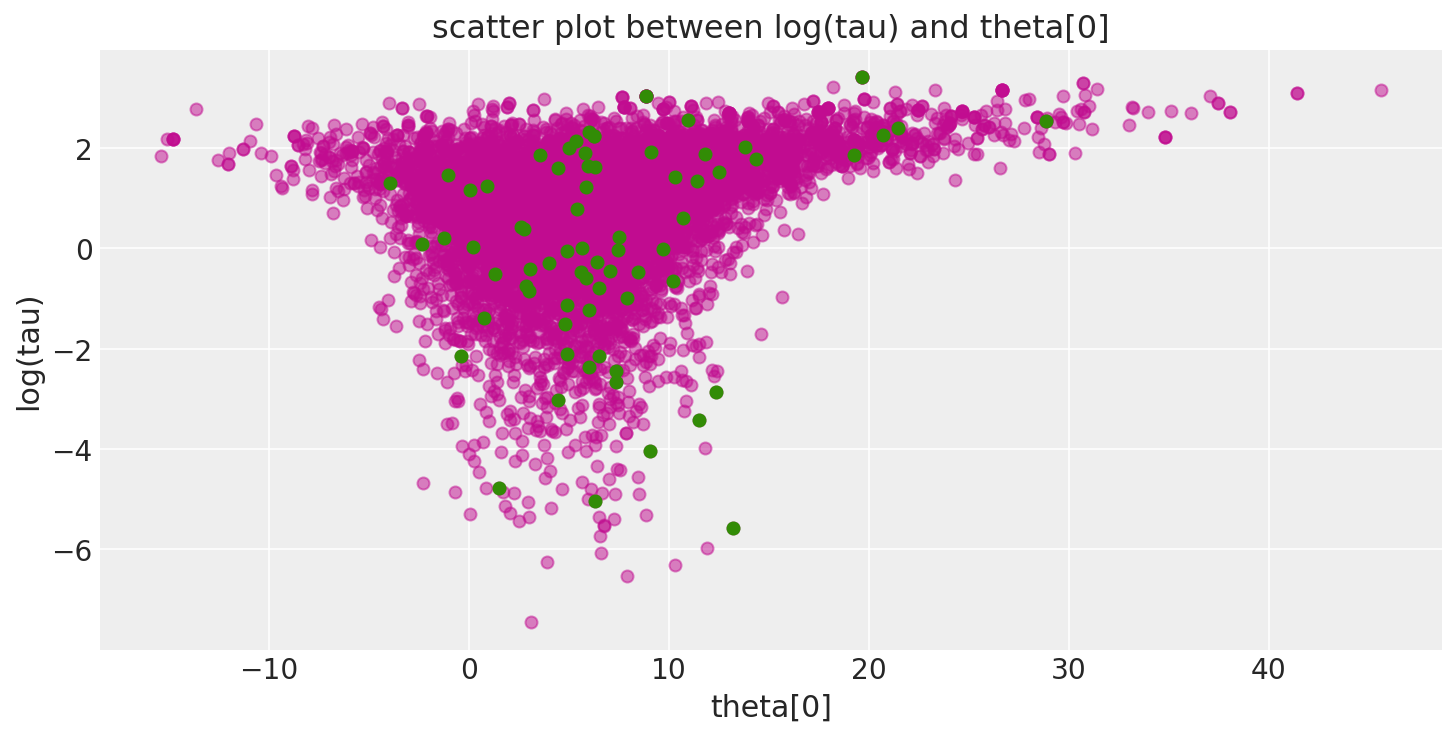
此外,由于发散跃迁(此处以绿色显示)倾向于位于病理附近,因此我们可以使用它们来识别参数空间中问题区域的位置。
def pairplot_divergence(trace, ax=None, divergence=True, color="C3", divergence_color="C2"):
theta = trace.get_values(varname="theta", combine=True)[:, 0]
logtau = trace.get_values(varname="tau_log__", combine=True)
if not ax:
_, ax = plt.subplots(1, 1, figsize=(10, 5))
ax.plot(theta, logtau, "o", color=color, alpha=0.5)
if divergence:
divergent = trace["diverging"]
ax.plot(theta[divergent], logtau[divergent], "o", color=divergence_color)
ax.set_xlabel("theta[0]")
ax.set_ylabel("log(tau)")
ax.set_title("scatter plot between log(tau) and theta[0]")
return ax
pairplot_divergence(short_trace);
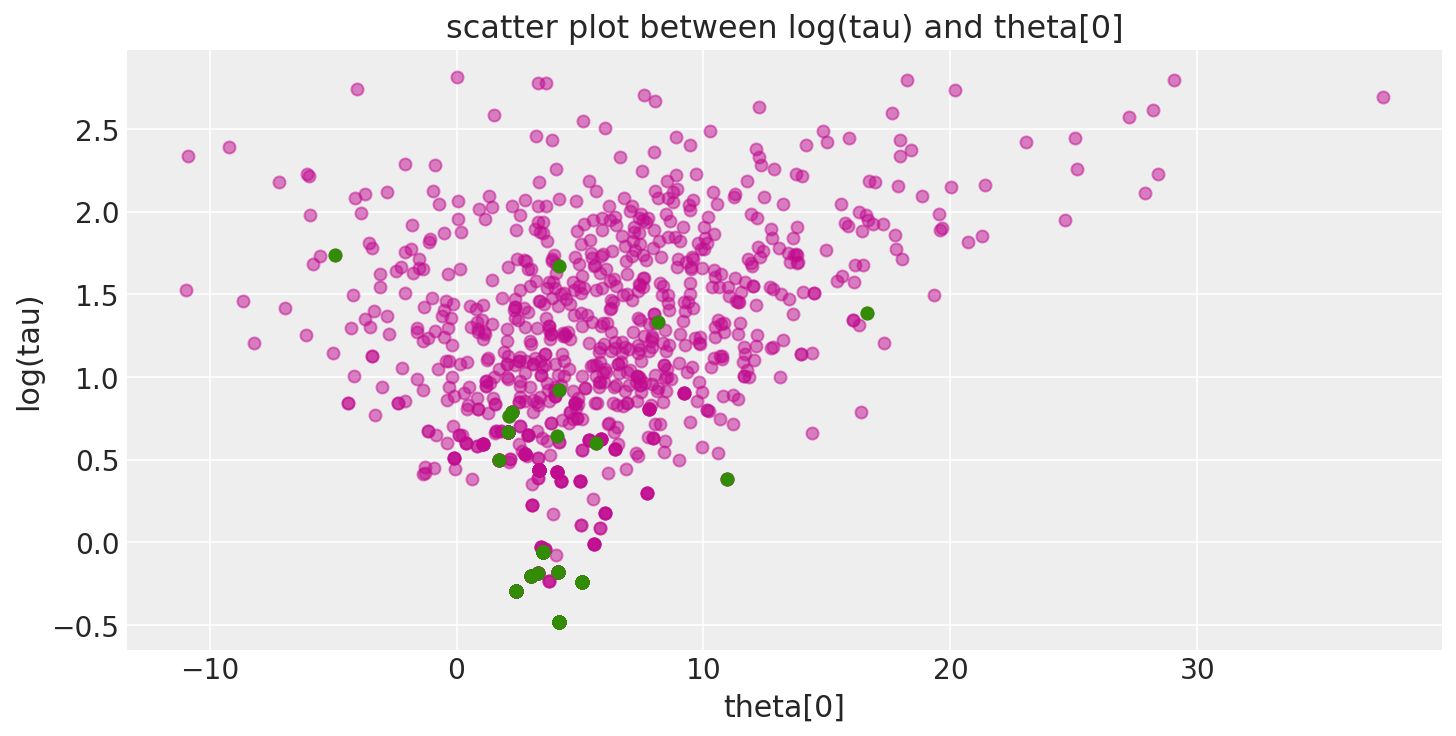
重要的是要指出,来自迹的病态样本不一定集中在漏斗处:当遇到发散时,正在构建的子树将被拒绝,并且跃迁会从现有的离散轨迹中均匀采样。因此,发散样本不会精确地位于高曲率区域。
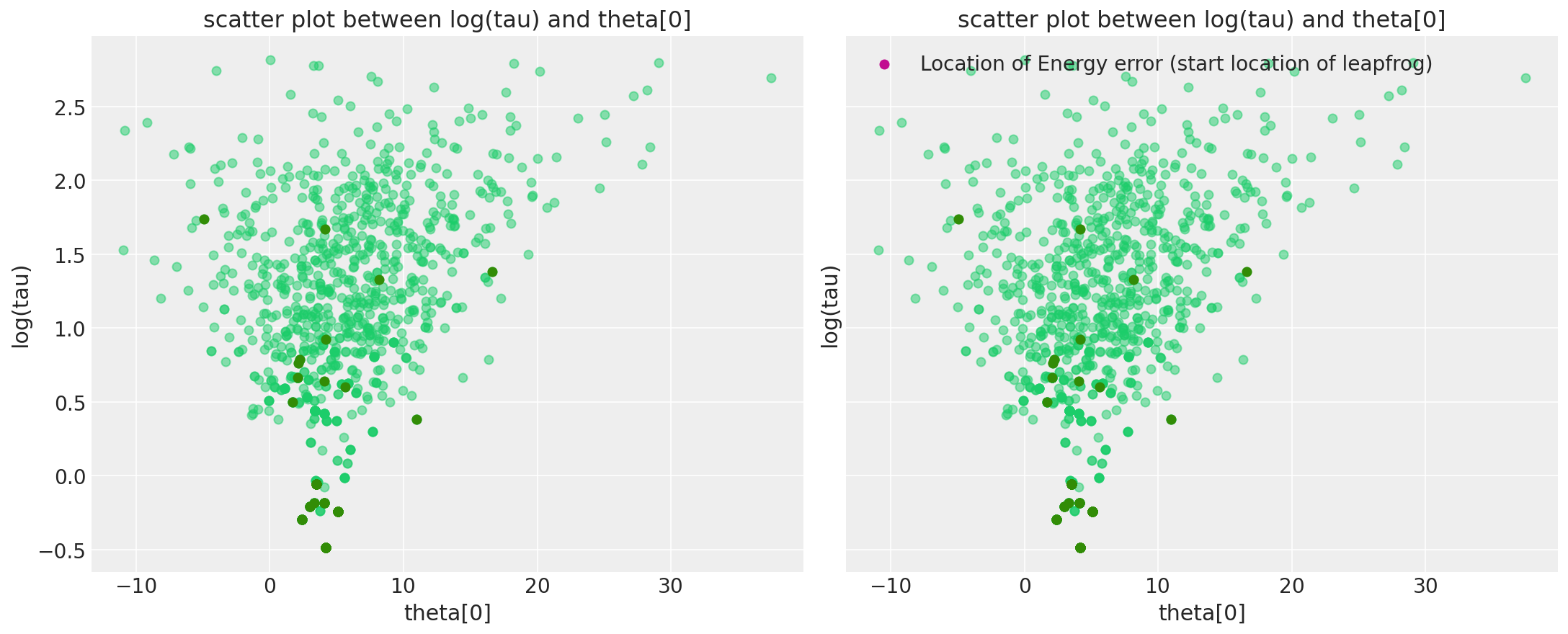
在 pymc3 中,我们最近实现了一个警告系统,该系统还保存了发散发生的位置的信息,因此您可以直接可视化它们。更准确地说,我们在警告中包含的发散点是该有问题的前跃步开始的点。有些也可能是因为发散发生在其中一个前跃步中(严格来说不是一个点)。但尽管如此,可视化这些应该可以更接近漏斗的位置。
请注意,仅存储前 100 个发散,这样我们就不会耗尽所有内存。
divergent_point = defaultdict(list)
chain_warn = short_trace.report._chain_warnings
for i in range(len(chain_warn)):
for warning_ in chain_warn[i]:
if warning_.step is not None and warning_.extra is not None:
for RV in Centered_eight.free_RVs:
para_name = RV.name
divergent_point[para_name].append(warning_.extra[para_name])
for RV in Centered_eight.free_RVs:
para_name = RV.name
divergent_point[para_name] = np.asarray(divergent_point[para_name])
tau_log_d = divergent_point["tau_log__"]
theta0_d = divergent_point["theta"]
Ndiv_recorded = len(tau_log_d)
_, ax = plt.subplots(1, 2, figsize=(15, 6), sharex=True, sharey=True)
pairplot_divergence(short_trace, ax=ax[0], color="C7", divergence_color="C2")
plt.title("scatter plot between log(tau) and theta[0]")
pairplot_divergence(short_trace, ax=ax[1], color="C7", divergence_color="C2")
theta_trace = short_trace["theta"]
theta0 = theta_trace[:, 0]
ax[1].plot(
[theta0[divergent == 1][:Ndiv_recorded], theta0_d],
[logtau[divergent == 1][:Ndiv_recorded], tau_log_d],
"k-",
alpha=0.5,
)
ax[1].scatter(
theta0_d, tau_log_d, color="C3", label="Location of Energy error (start location of leapfrog)"
)
plt.title("scatter plot between log(tau) and theta[0]")
plt.legend();
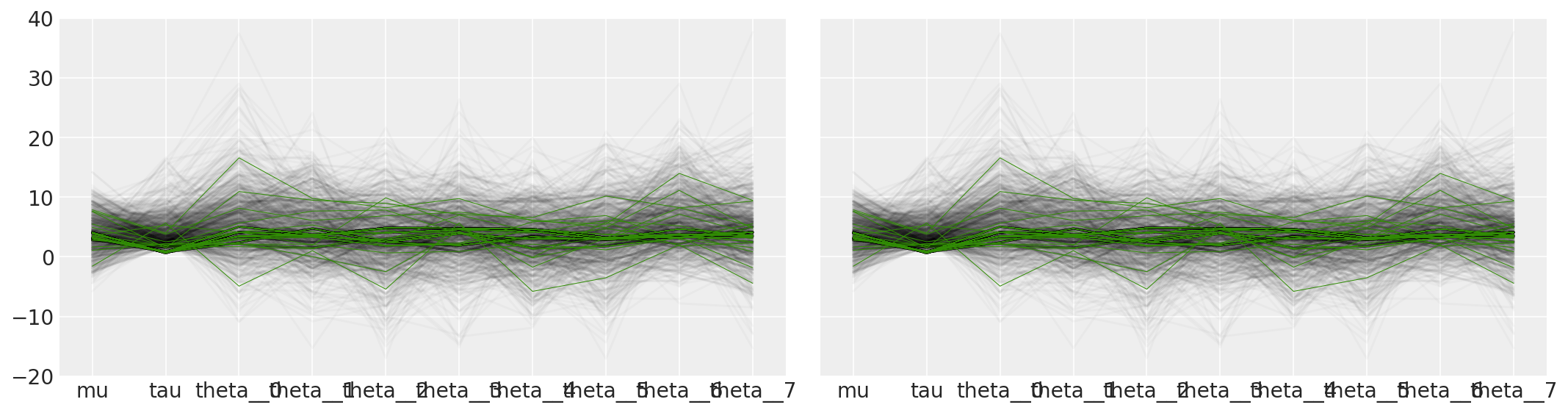
还有许多其他方法可以探索和可视化参数空间中的病态区域。例如,我们可以重现 贝叶斯工作流程中的可视化 中的图 5b
tracedf = pm.trace_to_dataframe(short_trace)
plotorder = [
"mu",
"tau",
"theta__0",
"theta__1",
"theta__2",
"theta__3",
"theta__4",
"theta__5",
"theta__6",
"theta__7",
]
tracedf = tracedf[plotorder]
_, ax = plt.subplots(1, 2, figsize=(15, 4), sharex=True, sharey=True)
ax[0].plot(tracedf.values[divergent == 0].T, color="k", alpha=0.025)
ax[0].plot(tracedf.values[divergent == 1].T, color="C2", lw=0.5)
ax[1].plot(tracedf.values[divergent == 0].T, color="k", alpha=0.025)
ax[1].plot(tracedf.values[divergent == 1].T, color="C2", lw=0.5)
divsp = np.hstack(
[
divergent_point["mu"],
np.exp(divergent_point["tau_log__"]),
divergent_point["theta"],
]
)
ax[1].plot(divsp.T, "C3", lw=0.5)
plt.ylim([-20, 40])
plt.xticks(range(10), plotorder)
plt.tight_layout()
/var/folders/f5/4hllfzqx6pq2sfm22_khf5400000gn/T/ipykernel_63426/2369948333.py:32: UserWarning: This figure was using constrained_layout, but that is incompatible with subplots_adjust and/or tight_layout; disabling constrained_layout.
plt.tight_layout()
# A small wrapper function for displaying the MCMC sampler diagnostics as above
def report_trace(trace):
# plot the trace of log(tau)
az.plot_trace({"log(tau)": trace.get_values(varname="tau_log__", combine=False)})
# plot the estimate for the mean of log(τ) cumulating mean
logtau = np.log(trace["tau"])
mlogtau = [np.mean(logtau[:i]) for i in np.arange(1, len(logtau))]
plt.figure(figsize=(15, 4))
plt.axhline(0.7657852, lw=2.5, color="gray")
plt.plot(mlogtau, lw=2.5)
plt.ylim(0, 2)
plt.xlabel("Iteration")
plt.ylabel("MCMC mean of log(tau)")
plt.title("MCMC estimation of log(tau)")
plt.show()
# display the total number and percentage of divergent
divergent = trace["diverging"]
print("Number of Divergent %d" % divergent.nonzero()[0].size)
divperc = divergent.nonzero()[0].size / len(trace) * 100
print("Percentage of Divergent %.1f" % divperc)
# scatter plot between log(tau) and theta[0]
# for the identification of the problematic neighborhoods in parameter space
pairplot_divergence(trace);
更安全、更长的马尔可夫链#
鉴于拆分 \(\hat{R}\) 在单个短链上的潜在不敏感性,Stan 建议始终运行多个链,时间尽可能长,以便有最佳机会观察到对几何遍历性的任何阻碍。但是,由于并非总是可以为复杂模型运行长链,因此发散是用于有偏 MCMC 估计的非常强大的诊断工具。
with Centered_eight:
longer_trace = pm.sample(4000, chains=2, tune=1000, random_seed=SEED)
/Users/reshamashaikh/miniforge3/envs/pymc-ex/lib/python3.10/site-packages/deprecat/classic.py:215: FutureWarning: In v4.0, pm.sample will return an `arviz.InferenceData` object instead of a `MultiTrace` by default. You can pass return_inferencedata=True or return_inferencedata=False to be safe and silence this warning.
return wrapped_(*args_, **kwargs_)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [theta, tau, mu]
Sampling 2 chains for 1_000 tune and 4_000 draw iterations (2_000 + 8_000 draws total) took 56 seconds.
There were 224 divergences after tuning. Increase `target_accept` or reparameterize.
The acceptance probability does not match the target. It is 0.5963528759316614, but should be close to 0.8. Try to increase the number of tuning steps.
There were 66 divergences after tuning. Increase `target_accept` or reparameterize.
The acceptance probability does not match the target. It is 0.614889465736071, but should be close to 0.8. Try to increase the number of tuning steps.
The rhat statistic is larger than 1.05 for some parameters. This indicates slight problems during sampling.
The estimated number of effective samples is smaller than 200 for some parameters.
report_trace(longer_trace)
Number of Divergent 290
Percentage of Divergent 7.2
az.summary(longer_trace).round(2)
Got error No model on context stack. trying to find log_likelihood in translation.
/Users/reshamashaikh/miniforge3/envs/pymc-ex/lib/python3.10/site-packages/arviz/data/io_pymc3_3x.py:98: FutureWarning: Using `from_pymc3` without the model will be deprecated in a future release. Not using the model will return less accurate and less useful results. Make sure you use the model argument or call from_pymc3 within a model context.
warnings.warn(
| 均值 | 标准差 | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| mu | 4.45 | 3.20 | -1.30 | 10.52 | 0.25 | 0.22 | 172.0 | 1723.0 | 1.01 |
| theta[0] | 6.42 | 5.63 | -2.97 | 18.08 | 0.20 | 0.14 | 497.0 | 2540.0 | 1.00 |
| theta[1] | 4.99 | 4.66 | -4.54 | 13.45 | 0.24 | 0.17 | 339.0 | 2300.0 | 1.01 |
| theta[2] | 3.97 | 5.33 | -6.64 | 13.66 | 0.25 | 0.18 | 302.0 | 2460.0 | 1.01 |
| theta[3] | 4.71 | 4.73 | -4.72 | 13.63 | 0.21 | 0.15 | 385.0 | 2574.0 | 1.01 |
| theta[4] | 3.65 | 4.60 | -5.26 | 12.23 | 0.26 | 0.18 | 272.0 | 2497.0 | 1.01 |
| theta[5] | 4.06 | 4.91 | -5.93 | 12.93 | 0.26 | 0.19 | 290.0 | 2266.0 | 1.00 |
| theta[6] | 6.36 | 4.96 | -1.99 | 16.76 | 0.15 | 0.10 | 771.0 | 2263.0 | 1.00 |
| theta[7] | 4.88 | 5.25 | -5.08 | 14.84 | 0.19 | 0.14 | 472.0 | 2634.0 | 1.01 |
| tau | 3.83 | 3.10 | 0.62 | 9.44 | 0.32 | 0.23 | 29.0 | 61.0 | 1.07 |
与 Stan 中的结果类似,\(\hat{R}\) 未指示任何严重问题。但是,每次迭代的有效样本量已大幅下降,表明我们运行时间越长,探索效率越低。这种奇怪的行为清楚地表明存在一些问题。如迹图所示,当链接近 \(\tau\) 的小值时,它偶尔会“粘滞”,这正是我们看到发散集中的地方。这清楚地表明了潜在的病理。这些粘滞间隔会在早期引起 MCMC 估计量的严重振荡,直到它们似乎最终稳定在有偏差的值中。
事实上,粘滞间隔是马尔可夫链试图纠正有偏差的探索。如果我们运行链的时间更长,那么它最终会再次卡住,并将 MCMC 估计量拉低到真实值。给定无限次迭代,这种微妙的平衡渐近于真实期望,正如我们根据 MCMC 的一致性保证所期望的那样。但是,在任何有限次迭代后停止都会破坏这种平衡,并使我们产生明显的偏差。
更多详细信息可以在 Betancourt 的 最新论文 中找到。
通过调整 PyMC3 的自适应例程来缓解发散#
哈密顿蒙特卡罗中的发散发生在哈密顿跃迁遇到曲率极大的区域时,例如分层漏斗的开口。由于无法准确解决这些区域,跃迁会发生故障并飞向无穷大。由于跃迁无法完全探索这些曲率极大的区域,因此我们失去了几何遍历性,并且我们的 MCMC 估计量变得有偏差。
Stan 中实现的算法使用启发式方法来快速识别这些行为不端的轨迹,从而标记发散,而无需等待它们一直运行到无穷大。但是,这种启发式方法可能有点激进,有时会将跃迁标记为发散,即使我们没有失去几何遍历性。
为了消除这种潜在的歧义,我们可以调整哈密顿跃迁的步长 \(\epsilon\)。步长越小,轨迹越准确,并且越不可能被错误标记为发散。换句话说,如果我们在哈密顿跃迁和目标分布之间具有几何遍历性,那么减小步长将减少,然后最终完全消除发散。但是,如果我们没有几何遍历性,那么减小步长将不会完全消除发散。
与 Stan 一样,PyMC3 中的步长在预热期间自动调整,但是我们可以通过调整 PyMC3 的自适应例程的配置来强制使用更小的步长。特别是,我们可以将 target_accept 参数从其默认值 0.8 增加到更接近其最大值 1。
调整自适应例程#
with Centered_eight:
fit_cp85 = pm.sample(5000, chains=2, tune=2000, target_accept=0.85)
/Users/reshamashaikh/miniforge3/envs/pymc-ex/lib/python3.10/site-packages/deprecat/classic.py:215: FutureWarning: In v4.0, pm.sample will return an `arviz.InferenceData` object instead of a `MultiTrace` by default. You can pass return_inferencedata=True or return_inferencedata=False to be safe and silence this warning.
return wrapped_(*args_, **kwargs_)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [theta, tau, mu]
Sampling 2 chains for 2_000 tune and 5_000 draw iterations (4_000 + 10_000 draws total) took 84 seconds.
There were 547 divergences after tuning. Increase `target_accept` or reparameterize.
The acceptance probability does not match the target. It is 0.4842846814954639, but should be close to 0.85. Try to increase the number of tuning steps.
There were 85 divergences after tuning. Increase `target_accept` or reparameterize.
The acceptance probability does not match the target. It is 0.737175456745239, but should be close to 0.85. Try to increase the number of tuning steps.
The rhat statistic is larger than 1.05 for some parameters. This indicates slight problems during sampling.
The estimated number of effective samples is smaller than 200 for some parameters.
with Centered_eight:
fit_cp90 = pm.sample(5000, chains=2, tune=2000, target_accept=0.90)
/Users/reshamashaikh/miniforge3/envs/pymc-ex/lib/python3.10/site-packages/deprecat/classic.py:215: FutureWarning: In v4.0, pm.sample will return an `arviz.InferenceData` object instead of a `MultiTrace` by default. You can pass return_inferencedata=True or return_inferencedata=False to be safe and silence this warning.
return wrapped_(*args_, **kwargs_)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [theta, tau, mu]
Sampling 2 chains for 2_000 tune and 5_000 draw iterations (4_000 + 10_000 draws total) took 91 seconds.
There were 430 divergences after tuning. Increase `target_accept` or reparameterize.
The acceptance probability does not match the target. It is 0.705290719027636, but should be close to 0.9. Try to increase the number of tuning steps.
There were 74 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.05 for some parameters. This indicates slight problems during sampling.
The estimated number of effective samples is smaller than 200 for some parameters.
with Centered_eight:
fit_cp95 = pm.sample(5000, chains=2, tune=2000, target_accept=0.95)
/Users/reshamashaikh/miniforge3/envs/pymc-ex/lib/python3.10/site-packages/deprecat/classic.py:215: FutureWarning: In v4.0, pm.sample will return an `arviz.InferenceData` object instead of a `MultiTrace` by default. You can pass return_inferencedata=True or return_inferencedata=False to be safe and silence this warning.
return wrapped_(*args_, **kwargs_)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [theta, tau, mu]
Sampling 2 chains for 2_000 tune and 5_000 draw iterations (4_000 + 10_000 draws total) took 129 seconds.
There were 219 divergences after tuning. Increase `target_accept` or reparameterize.
The acceptance probability does not match the target. It is 0.8819302505195916, but should be close to 0.95. Try to increase the number of tuning steps.
There were 43 divergences after tuning. Increase `target_accept` or reparameterize.
The number of effective samples is smaller than 10% for some parameters.
with Centered_eight:
fit_cp99 = pm.sample(5000, chains=2, tune=2000, target_accept=0.99)
/Users/reshamashaikh/miniforge3/envs/pymc-ex/lib/python3.10/site-packages/deprecat/classic.py:215: FutureWarning: In v4.0, pm.sample will return an `arviz.InferenceData` object instead of a `MultiTrace` by default. You can pass return_inferencedata=True or return_inferencedata=False to be safe and silence this warning.
return wrapped_(*args_, **kwargs_)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [theta, tau, mu]
Sampling 2 chains for 2_000 tune and 5_000 draw iterations (4_000 + 10_000 draws total) took 227 seconds.
There were 40 divergences after tuning. Increase `target_accept` or reparameterize.
The acceptance probability does not match the target. It is 0.9693984517210503, but should be close to 0.99. Try to increase the number of tuning steps.
There were 7 divergences after tuning. Increase `target_accept` or reparameterize.
The number of effective samples is smaller than 10% for some parameters.
df = pd.DataFrame(
[
longer_trace["step_size"].mean(),
fit_cp85["step_size"].mean(),
fit_cp90["step_size"].mean(),
fit_cp95["step_size"].mean(),
fit_cp99["step_size"].mean(),
],
columns=["Step_size"],
)
df["Divergent"] = pd.Series(
[
longer_trace["diverging"].sum(),
fit_cp85["diverging"].sum(),
fit_cp90["diverging"].sum(),
fit_cp95["diverging"].sum(),
fit_cp99["diverging"].sum(),
]
)
df["delta_target"] = pd.Series([".80", ".85", ".90", ".95", ".99"])
df
| Step_size | 发散 | delta_target | |
|---|---|---|---|
| 0 | 0.276504 | 290 | .80 |
| 1 | 0.244083 | 632 | .85 |
| 2 | 0.164192 | 504 | .90 |
| 3 | 0.137629 | 262 | .95 |
| 4 | 0.043080 | 47 | .99 |
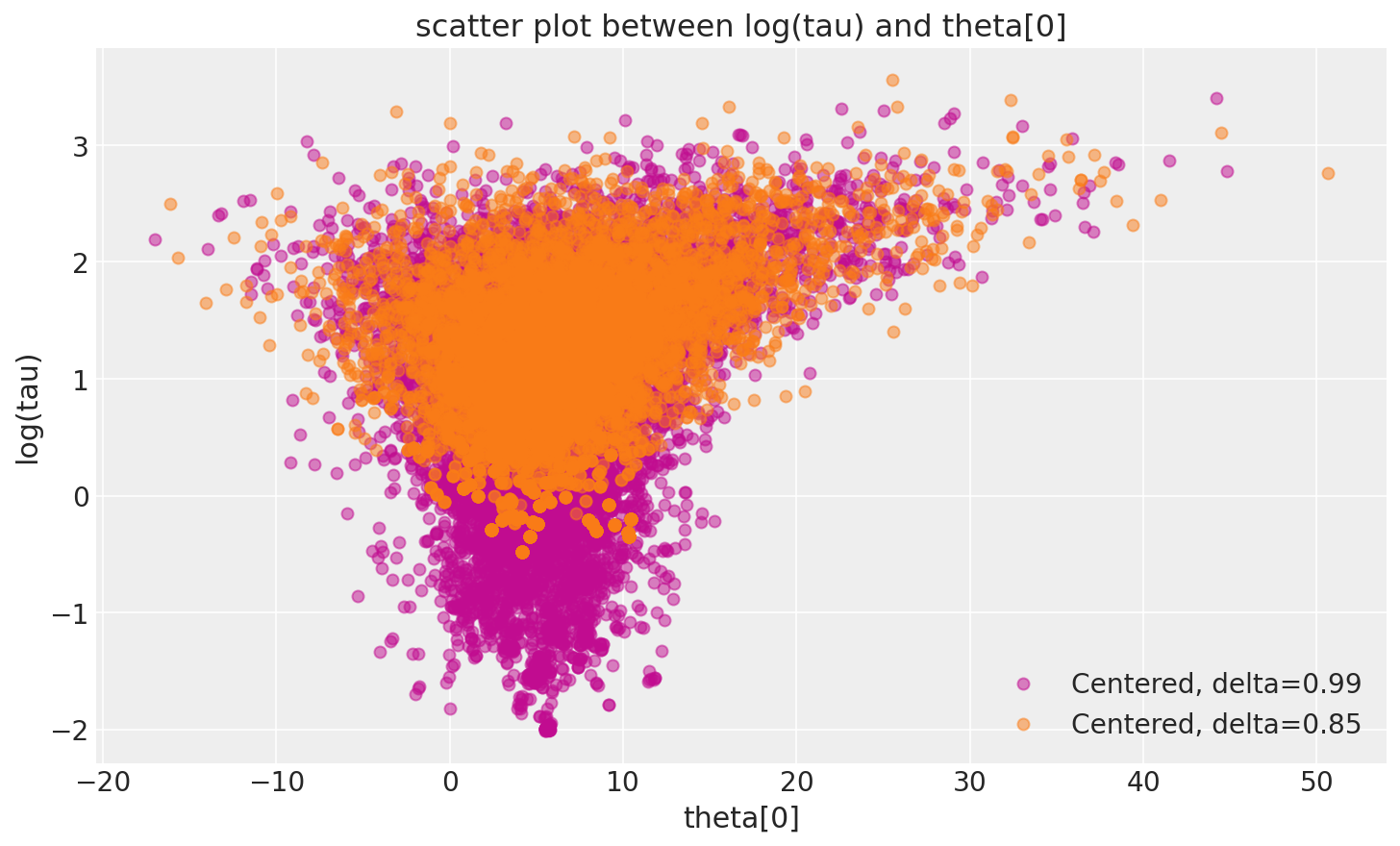
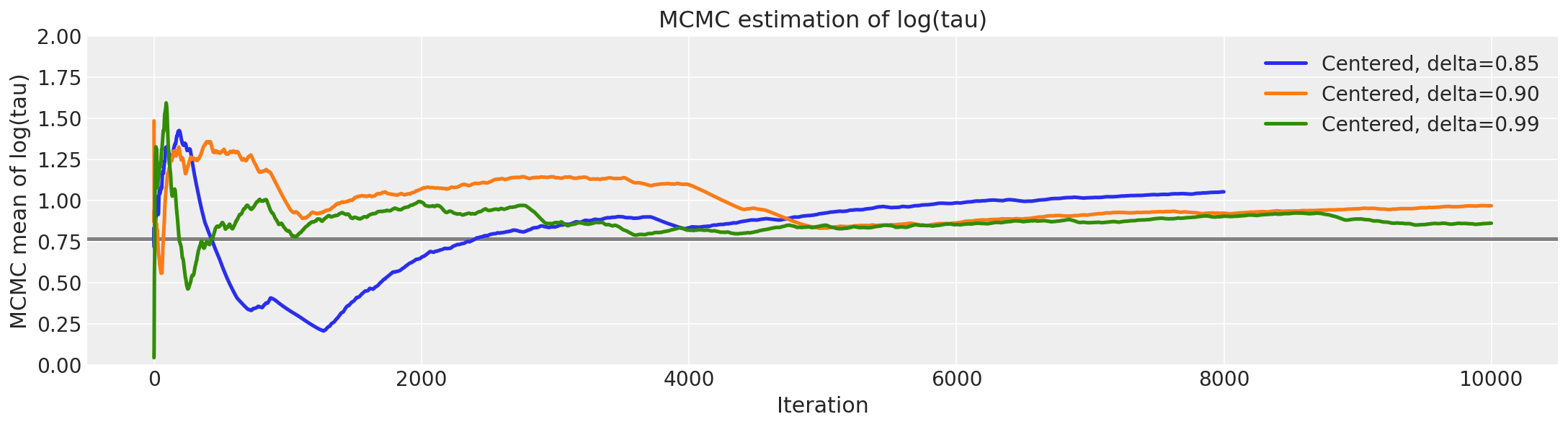
在此,当 delta 增加到 0.99 时,发散跃迁的数量急剧下降。
这种行为也具有很好的几何直觉。我们越减小步长,哈密顿马尔可夫链就越能探索漏斗的颈部。因此,随着步长的减小,\(log (\tau)\) 的边际后验分布会进一步延伸到负值。
由于在 PyMC3 中,调整后我们得到的步长比 Stan 小,因此几何形状得到了更好的探索。
但是,哈密顿跃迁仍然不关于八所学校模型的中心化实现是几何遍历的。事实上,鉴于观察到的偏差,这是预期的。
_, ax = plt.subplots(1, 1, figsize=(10, 6))
pairplot_divergence(fit_cp99, ax=ax, color="C3", divergence=False)
pairplot_divergence(longer_trace, ax=ax, color="C1", divergence=False)
ax.legend(["Centered, delta=0.99", "Centered, delta=0.85"]);
logtau0 = longer_trace["tau_log__"]
logtau2 = np.log(fit_cp90["tau"])
logtau1 = fit_cp99["tau_log__"]
plt.figure(figsize=(15, 4))
plt.axhline(0.7657852, lw=2.5, color="gray")
mlogtau0 = [np.mean(logtau0[:i]) for i in np.arange(1, len(logtau0))]
plt.plot(mlogtau0, label="Centered, delta=0.85", lw=2.5)
mlogtau2 = [np.mean(logtau2[:i]) for i in np.arange(1, len(logtau2))]
plt.plot(mlogtau2, label="Centered, delta=0.90", lw=2.5)
mlogtau1 = [np.mean(logtau1[:i]) for i in np.arange(1, len(logtau1))]
plt.plot(mlogtau1, label="Centered, delta=0.99", lw=2.5)
plt.ylim(0, 2)
plt.xlabel("Iteration")
plt.ylabel("MCMC mean of log(tau)")
plt.title("MCMC estimation of log(tau)")
plt.legend();
非中心化八所学校实施#
虽然减小步长可以改善探索,但最终它只会揭示中心化实现中病理的真实程度。幸运的是,还有另一种实现分层模型的方法,它不会遭受相同的病理。
在非中心化参数化中,我们不尝试直接拟合组级参数,而是拟合一个潜在高斯变量,我们可以从中通过缩放和平移来恢复组级参数。
Stan 模型
data {
int<lower=0> J;
real y[J];
real<lower=0> sigma[J];
}
parameters {
real mu;
real<lower=0> tau;
real theta_tilde[J];
}
transformed parameters {
real theta[J];
for (j in 1:J)
theta[j] = mu + tau * theta_tilde[j];
}
model {
mu ~ normal(0, 5);
tau ~ cauchy(0, 5);
theta_tilde ~ normal(0, 1);
y ~ normal(theta, sigma);
}
with pm.Model() as NonCentered_eight:
mu = pm.Normal("mu", mu=0, sigma=5)
tau = pm.HalfCauchy("tau", beta=5)
theta_tilde = pm.Normal("theta_t", mu=0, sigma=1, shape=J)
theta = pm.Deterministic("theta", mu + tau * theta_tilde)
obs = pm.Normal("obs", mu=theta, sigma=sigma, observed=y)
with NonCentered_eight:
fit_ncp80 = pm.sample(5000, chains=2, tune=1000, random_seed=SEED, target_accept=0.80)
/Users/reshamashaikh/miniforge3/envs/pymc-ex/lib/python3.10/site-packages/deprecat/classic.py:215: FutureWarning: In v4.0, pm.sample will return an `arviz.InferenceData` object instead of a `MultiTrace` by default. You can pass return_inferencedata=True or return_inferencedata=False to be safe and silence this warning.
return wrapped_(*args_, **kwargs_)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [theta_t, tau, mu]
Sampling 2 chains for 1_000 tune and 5_000 draw iterations (2_000 + 10_000 draws total) took 32 seconds.
There were 19 divergences after tuning. Increase `target_accept` or reparameterize.
There were 52 divergences after tuning. Increase `target_accept` or reparameterize.
az.summary(fit_ncp80).round(2)
Got error No model on context stack. trying to find log_likelihood in translation.
/Users/reshamashaikh/miniforge3/envs/pymc-ex/lib/python3.10/site-packages/arviz/data/io_pymc3_3x.py:98: FutureWarning: Using `from_pymc3` without the model will be deprecated in a future release. Not using the model will return less accurate and less useful results. Make sure you use the model argument or call from_pymc3 within a model context.
warnings.warn(
| 均值 | 标准差 | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| mu | 4.39 | 3.29 | -1.82 | 10.48 | 0.04 | 0.03 | 7993.0 | 4425.0 | 1.0 |
| theta_t[0] | 0.32 | 0.97 | -1.44 | 2.19 | 0.01 | 0.01 | 8723.0 | 5684.0 | 1.0 |
| theta_t[1] | 0.10 | 0.94 | -1.66 | 1.84 | 0.01 | 0.01 | 10767.0 | 6229.0 | 1.0 |
| theta_t[2] | -0.10 | 0.96 | -1.94 | 1.68 | 0.01 | 0.01 | 9773.0 | 5893.0 | 1.0 |
| theta_t[3] | 0.08 | 0.95 | -1.75 | 1.83 | 0.01 | 0.01 | 10138.0 | 6101.0 | 1.0 |
| theta_t[4] | -0.17 | 0.92 | -1.91 | 1.60 | 0.01 | 0.01 | 8721.0 | 6476.0 | 1.0 |
| theta_t[5] | -0.07 | 0.94 | -1.85 | 1.67 | 0.01 | 0.01 | 11379.0 | 7066.0 | 1.0 |
| theta_t[6] | 0.36 | 0.96 | -1.47 | 2.13 | 0.01 | 0.01 | 9317.0 | 6189.0 | 1.0 |
| theta_t[7] | 0.07 | 0.98 | -1.72 | 1.94 | 0.01 | 0.01 | 11444.0 | 6889.0 | 1.0 |
| tau | 3.64 | 3.36 | 0.00 | 9.39 | 0.05 | 0.04 | 4430.0 | 3569.0 | 1.0 |
| theta[0] | 6.26 | 5.57 | -4.45 | 16.36 | 0.07 | 0.06 | 6821.0 | 4801.0 | 1.0 |
| theta[1] | 4.93 | 4.55 | -3.61 | 13.80 | 0.05 | 0.04 | 9825.0 | 6967.0 | 1.0 |
| theta[2] | 3.84 | 5.30 | -5.75 | 14.24 | 0.07 | 0.06 | 7421.0 | 5379.0 | 1.0 |
| theta[3] | 4.86 | 4.85 | -3.93 | 14.24 | 0.05 | 0.05 | 8766.0 | 6023.0 | 1.0 |
| theta[4] | 3.57 | 4.64 | -5.70 | 11.97 | 0.05 | 0.04 | 8191.0 | 5926.0 | 1.0 |
| theta[5] | 4.02 | 4.90 | -4.93 | 13.28 | 0.06 | 0.05 | 7713.0 | 6105.0 | 1.0 |
| theta[6] | 6.35 | 4.99 | -2.62 | 16.06 | 0.06 | 0.04 | 8799.0 | 5610.0 | 1.0 |
| theta[7] | 4.92 | 5.33 | -4.54 | 15.72 | 0.06 | 0.04 | 8565.0 | 6393.0 | 1.0 |
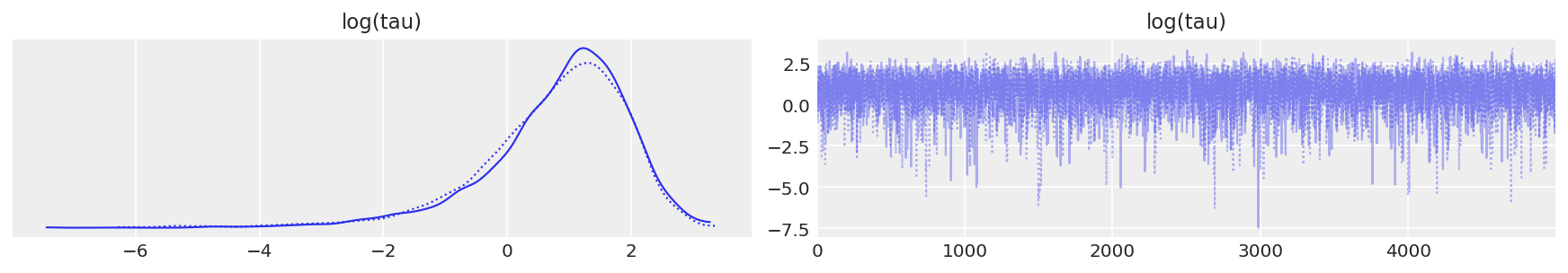
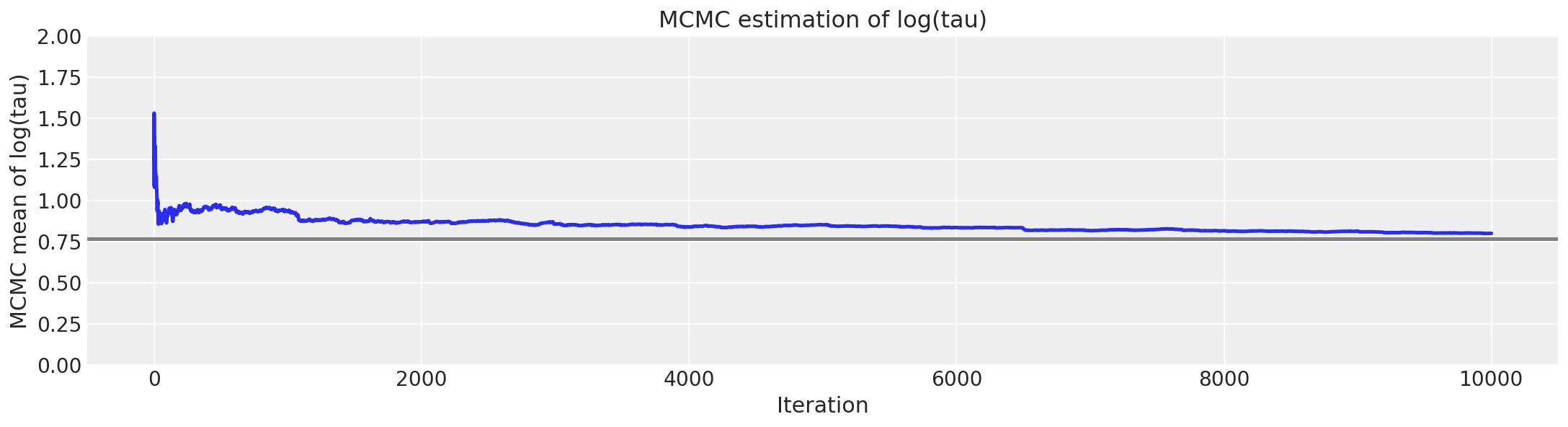
如上所示,每次迭代的有效样本量已大大提高,并且迹图不再显示任何“粘滞性”。但是,我们仍然看到罕见的发散。这些不频繁的发散似乎没有集中在参数空间中的任何位置,这表明发散是误报。
report_trace(fit_ncp80)
Number of Divergent 71
Percentage of Divergent 1.4
正如对误报的预期一样,我们可以通过减小步长来完全消除发散。
with NonCentered_eight:
fit_ncp90 = pm.sample(5000, chains=2, tune=1000, random_seed=SEED, target_accept=0.90)
# display the total number and percentage of divergent
divergent = fit_ncp90["diverging"]
print("Number of Divergent %d" % divergent.nonzero()[0].size)
/Users/reshamashaikh/miniforge3/envs/pymc-ex/lib/python3.10/site-packages/deprecat/classic.py:215: FutureWarning: In v4.0, pm.sample will return an `arviz.InferenceData` object instead of a `MultiTrace` by default. You can pass return_inferencedata=True or return_inferencedata=False to be safe and silence this warning.
return wrapped_(*args_, **kwargs_)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [theta_t, tau, mu]
Sampling 2 chains for 1_000 tune and 5_000 draw iterations (2_000 + 10_000 draws total) took 35 seconds.
There was 1 divergence after tuning. Increase `target_accept` or reparameterize.
Number of Divergent 1
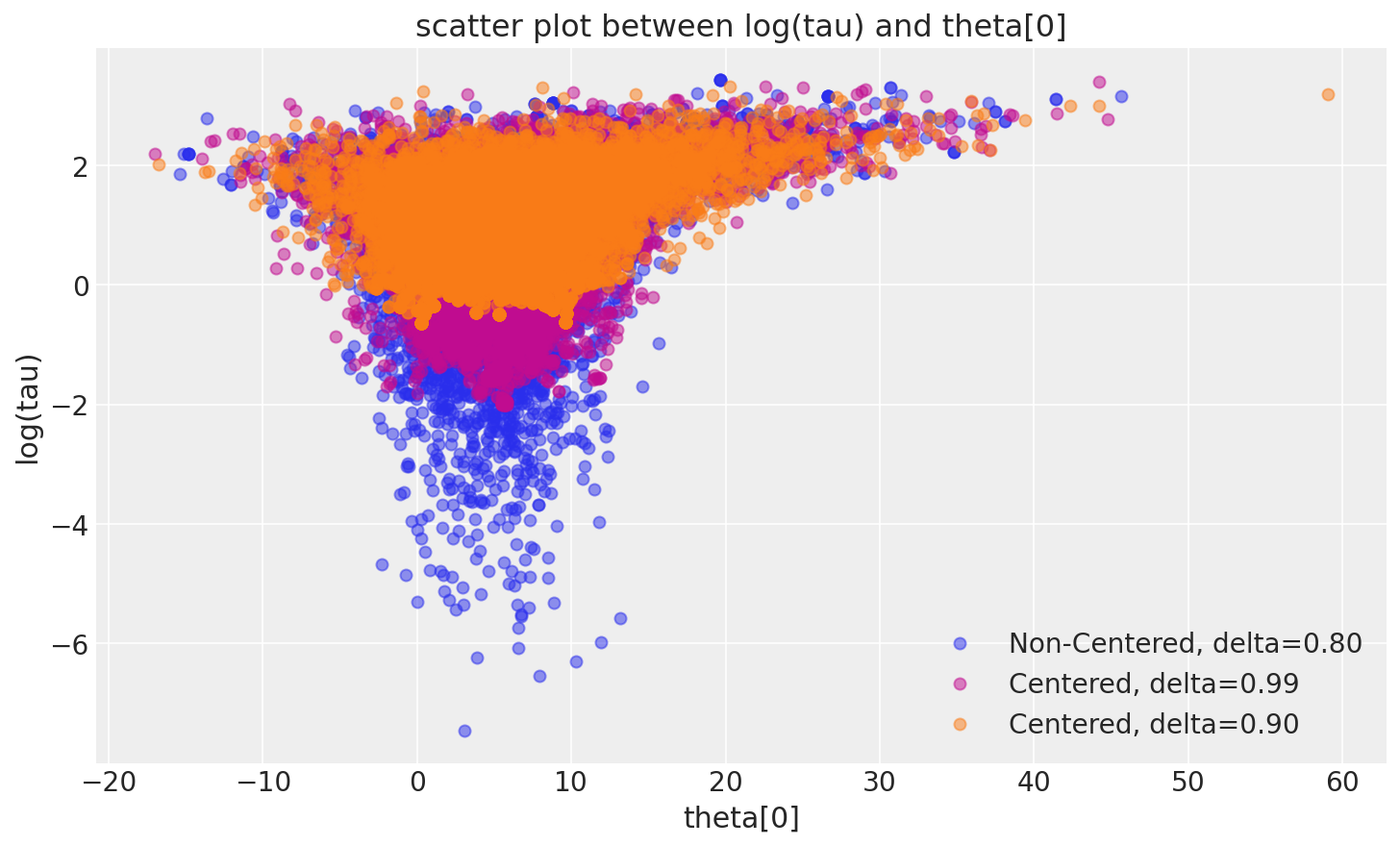
非中心化实现更令人愉悦的几何形状允许马尔可夫链深入探索漏斗的颈部,捕获与测量值一致的 tau (\(\tau\)) 的最小值。因此,来自非中心化链的 MCMC 估计量迅速收敛到其真实期望值。
_, ax = plt.subplots(1, 1, figsize=(10, 6))
pairplot_divergence(fit_ncp80, ax=ax, color="C0", divergence=False)
pairplot_divergence(fit_cp99, ax=ax, color="C3", divergence=False)
pairplot_divergence(fit_cp90, ax=ax, color="C1", divergence=False)
ax.legend(["Non-Centered, delta=0.80", "Centered, delta=0.99", "Centered, delta=0.90"]);
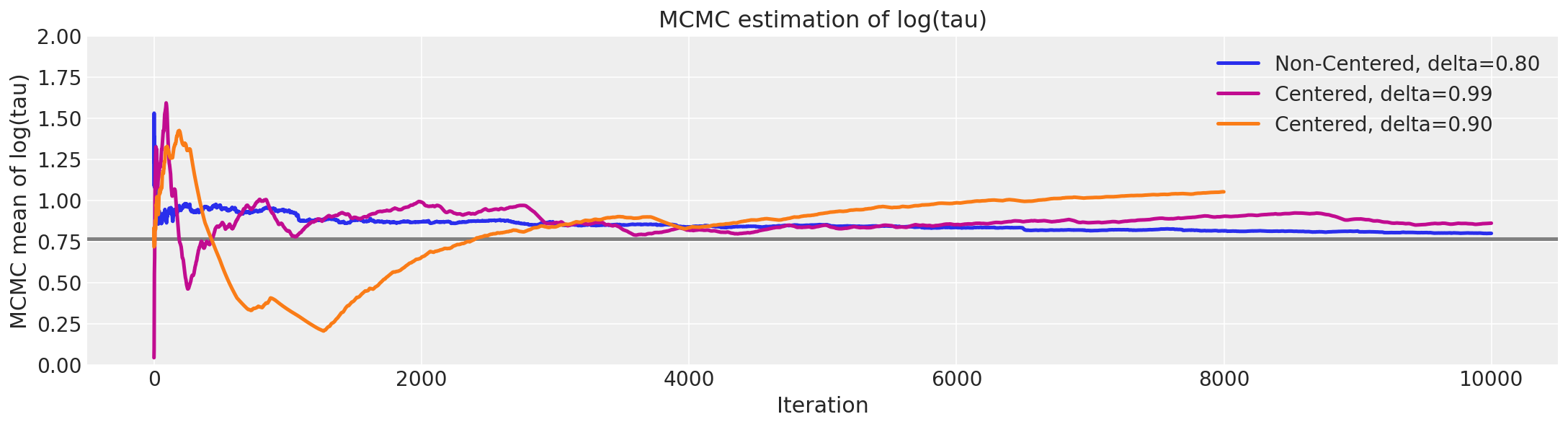
logtaun = fit_ncp80["tau_log__"]
plt.figure(figsize=(15, 4))
plt.axhline(0.7657852, lw=2.5, color="gray")
mlogtaun = [np.mean(logtaun[:i]) for i in np.arange(1, len(logtaun))]
plt.plot(mlogtaun, color="C0", lw=2.5, label="Non-Centered, delta=0.80")
mlogtau1 = [np.mean(logtau1[:i]) for i in np.arange(1, len(logtau1))]
plt.plot(mlogtau1, color="C3", lw=2.5, label="Centered, delta=0.99")
mlogtau0 = [np.mean(logtau0[:i]) for i in np.arange(1, len(logtau0))]
plt.plot(mlogtau0, color="C1", lw=2.5, label="Centered, delta=0.90")
plt.ylim(0, 2)
plt.xlabel("Iteration")
plt.ylabel("MCMC mean of log(tau)")
plt.title("MCMC estimation of log(tau)")
plt.legend();
许可证声明#
此示例 галерея 中的所有笔记本均根据 MIT 许可证 提供,该许可证允许修改和再分发以用于任何用途,前提是保留版权和许可证声明。
引用 PyMC 示例#
要引用此笔记本,请使用 Zenodo 为 pymc-examples 存储库提供的 DOI。
重要提示
许多笔记本都改编自其他来源:博客、书籍...... 在这种情况下,您也应该引用原始来源。
另请记住引用代码使用的相关库。
这是一个 bibtex 中的引用模板
@incollection{citekey,
author = "<notebook authors, see above>",
title = "<notebook title>",
editor = "PyMC Team",
booktitle = "PyMC examples",
doi = "10.5281/zenodo.5654871"
}
一旦渲染,它可能看起来像