边缘似然实现#
gp.Marginal 类实现了 GP 回归更常见的情况:观测数据是 GP 和高斯噪声的总和。gp.Marginal 具有 marginal_likelihood 方法、conditional 方法和 predict 方法。给定均值和协方差函数,函数 \(f(x)\) 被建模为,
观测值 \(y\) 是未知函数加上噪声
.marginal_likelihood 方法#
未知的潜在函数可以从 GP 先验概率与正态似然的乘积中解析积分出来。这个量被称为边缘似然。
边缘似然的对数,\(p(y \mid x)\),是
\(\boldsymbol\Sigma\) 是高斯噪声的协方差矩阵。由于高斯噪声不需要是白噪声才能共轭,因此当提供标量时,marginal_likelihood 方法支持使用白噪声项,或者当提供协方差函数时,支持使用噪声协方差函数。
gp.marginal_likelihood 方法实现了上面给出的量。一些示例代码如下,
import numpy as np
import pymc3 as pm
# A one dimensional column vector of inputs.
X = np.linspace(0, 1, 10)[:,None]
with pm.Model() as marginal_gp_model:
# Specify the covariance function.
cov_func = pm.gp.cov.ExpQuad(1, ls=0.1)
# Specify the GP. The default mean function is `Zero`.
gp = pm.gp.Marginal(cov_func=cov_func)
# The scale of the white noise term can be provided,
sigma = pm.HalfCauchy("sigma", beta=5)
y_ = gp.marginal_likelihood("y", X=X, y=y, sigma=sigma)
# OR a covariance function for the noise can be given
# noise_l = pm.Gamma("noise_l", alpha=2, beta=2)
# cov_func_noise = pm.gp.cov.Exponential(1, noise_l) + pm.gp.cov.WhiteNoise(sigma=0.1)
# y_ = gp.marginal_likelihood("y", X=X, y=y, sigma=cov_func_noise)
.conditional 分布#
.conditional 有一个可选标志 pred_noise,默认为 False。当 pred_sigma=False 时,conditional 方法生成 GP 表示的底层函数的预测分布。当 pred_sigma=True 时,conditional 方法生成 GP 加噪声的预测分布。使用上面定义的相同 gp 对象,
# vector of new X points we want to predict the function at
Xnew = np.linspace(0, 2, 100)[:, None]
with marginal_gp_model:
f_star = gp.conditional("f_star", Xnew=Xnew)
# or to predict the GP plus noise
y_star = gp.conditional("y_star", Xnew=Xnew, pred_sigma=True)
如果使用加性 GP 模型,可以通过设置可选参数 given 来构建各个分量的条件分布。有关构建加性 GP 的更多信息,请参阅主文档页面。有关示例,请参阅 Mauna Loa CO\(_2\) 笔记本。
进行预测#
.predict 方法返回给定 point 的 gp 的条件均值和方差作为 NumPy 数组。point 可以是 find_MAP 的结果或来自迹的样本。.predict 方法可以在 Model 块外部使用。与 .conditional 类似,.predict 接受 given,因此它可以从加性 GP 的分量生成预测。
# The mean and full covariance
mu, cov = gp.predict(Xnew, point=trace[-1])
# The mean and variance (diagonal of the covariance)
mu, var = gp.predict(Xnew, point=trace[-1], diag=True)
# With noise included
mu, var = gp.predict(Xnew, point=trace[-1], diag=True, pred_sigma=True)
示例:使用白色高斯噪声进行回归#
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pymc as pm
import scipy as sp
%matplotlib inline
# set the seed
np.random.seed(1)
n = 100 # The number of data points
X = np.linspace(0, 10, n)[:, None] # The inputs to the GP, they must be arranged as a column vector
# Define the true covariance function and its parameters
ell_true = 1.0
eta_true = 3.0
cov_func = eta_true**2 * pm.gp.cov.Matern52(1, ell_true)
# A mean function that is zero everywhere
mean_func = pm.gp.mean.Zero()
# The latent function values are one sample from a multivariate normal
# Note that we have to call `eval()` because PyMC3 built on top of Theano
f_true = np.random.multivariate_normal(
mean_func(X).eval(), cov_func(X).eval() + 1e-8 * np.eye(n), 1
).flatten()
# The observed data is the latent function plus a small amount of IID Gaussian noise
# The standard deviation of the noise is `sigma`
sigma_true = 2.0
y = f_true + sigma_true * np.random.randn(n)
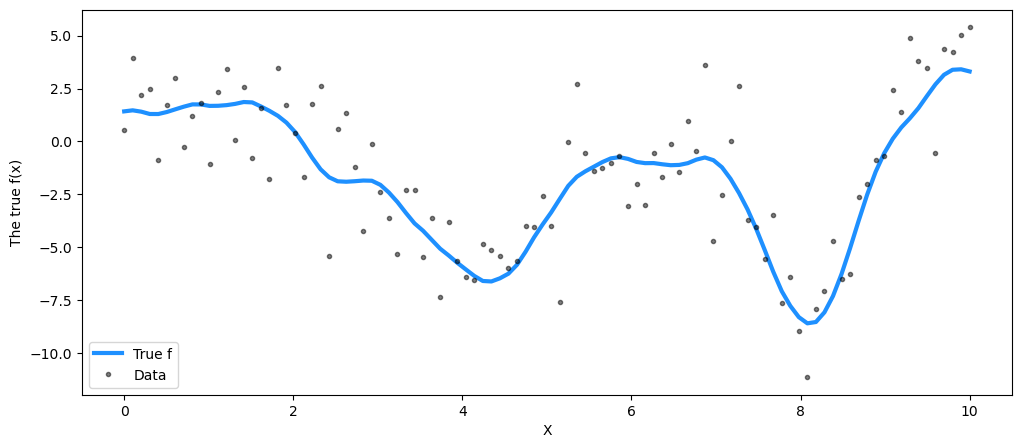
## Plot the data and the unobserved latent function
fig = plt.figure(figsize=(12, 5))
ax = fig.gca()
ax.plot(X, f_true, "dodgerblue", lw=3, label="True f")
ax.plot(X, y, "ok", ms=3, alpha=0.5, label="Data")
ax.set_xlabel("X")
ax.set_ylabel("The true f(x)")
plt.legend();
with pm.Model() as model:
ell = pm.Gamma("ell", alpha=2, beta=1)
eta = pm.HalfCauchy("eta", beta=5)
cov = eta**2 * pm.gp.cov.Matern52(1, ell)
gp = pm.gp.Marginal(cov_func=cov)
sigma = pm.HalfCauchy("sigma", beta=5)
y_ = gp.marginal_likelihood("y", X=X, y=y, sigma=sigma)
with model:
marginal_post = pm.sample(500, tune=2000, nuts_sampler="numpyro", chains=1)
/Users/cfonnesbeck/mambaforge/envs/bayes_course/lib/python3.11/site-packages/pymc/sampling/mcmc.py:243: UserWarning: Use of external NUTS sampler is still experimental
warnings.warn("Use of external NUTS sampler is still experimental", UserWarning)
Compiling...
Compilation time = 0:00:00.634832
Sampling...
sample: 100%|██████████| 2500/2500 [00:20<00:00, 119.94it/s, 7 steps of size 5.25e-01. acc. prob=0.92]
Sampling time = 0:00:23.334210
Transforming variables...
Transformation time = 0:00:00.014305
summary = az.summary(marginal_post, var_names=["ell", "eta", "sigma"], round_to=2, kind="stats")
summary["True value"] = [ell_true, eta_true, sigma_true]
summary
| 均值 | 标准差 | hdi_3% | hdi_97% | 真实值 | |
|---|---|---|---|---|---|
| ell | 1.18 | 0.28 | 0.72 | 1.76 | 1.0 |
| eta | 4.39 | 1.30 | 2.67 | 7.09 | 3.0 |
| sigma | 1.93 | 0.14 | 1.68 | 2.20 | 2.0 |
估计值接近它们的真实值。
使用 .conditional#
# new values from x=0 to x=20
X_new = np.linspace(0, 20, 600)[:, None]
# add the GP conditional to the model, given the new X values
with model:
f_pred = gp.conditional("f_pred", X_new)
with model:
pred_samples = pm.sample_posterior_predictive(
marginal_post.sel(draw=slice(0, 20)), var_names=["f_pred"]
)
Sampling: [f_pred]
# plot the results
fig = plt.figure(figsize=(12, 5))
ax = fig.gca()
# plot the samples from the gp posterior with samples and shading
from pymc.gp.util import plot_gp_dist
f_pred_samples = az.extract(pred_samples, group="posterior_predictive", var_names=["f_pred"])
plot_gp_dist(ax, samples=f_pred_samples.T, x=X_new)
# plot the data and the true latent function
plt.plot(X, f_true, "dodgerblue", lw=3, label="True f")
plt.plot(X, y, "ok", ms=3, alpha=0.5, label="Observed data")
# axis labels and title
plt.xlabel("X")
plt.ylim([-13, 13])
plt.title("Posterior distribution over $f(x)$ at the observed values")
plt.legend();
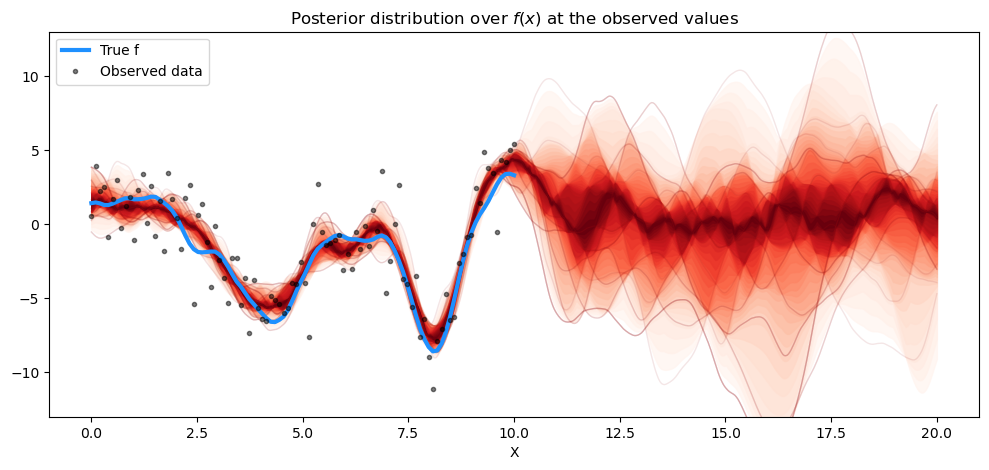
预测也与 gp.Latent 的结果非常吻合。预测新数据点怎么样?这里我们只预测了 \(f_*\),而不是 \(f_*\) + 噪声,这才是我们实际观察到的。
gp.Marginal 的 conditional 方法包含标志 pred_noise,其默认值为 False。要从后验预测分布中抽样,我们只需将此标志设置为 True。
with model:
y_pred = gp.conditional("y_pred", X_new, pred_noise=True)
y_samples = pm.sample_posterior_predictive(
marginal_post.sel(draw=slice(0, 50)), var_names=["y_pred"]
)
Sampling: [y_pred]
fig = plt.figure(figsize=(12, 5))
ax = fig.gca()
# posterior predictive distribution
y_pred_samples = az.extract(y_samples, group="posterior_predictive", var_names=["y_pred"])
plot_gp_dist(ax, y_pred_samples.T, X_new, plot_samples=False, palette="bone_r")
# overlay a scatter of one draw of random points from the
# posterior predictive distribution
plt.plot(X_new, y_pred_samples.sel(sample=1), "co", ms=2, label="Predicted data")
# plot original data and true function
plt.plot(X, y, "ok", ms=3, alpha=1.0, label="observed data")
plt.plot(X, f_true, "dodgerblue", lw=3, label="true f")
plt.xlabel("x")
plt.ylim([-13, 13])
plt.title("posterior predictive distribution, y_*")
plt.legend();
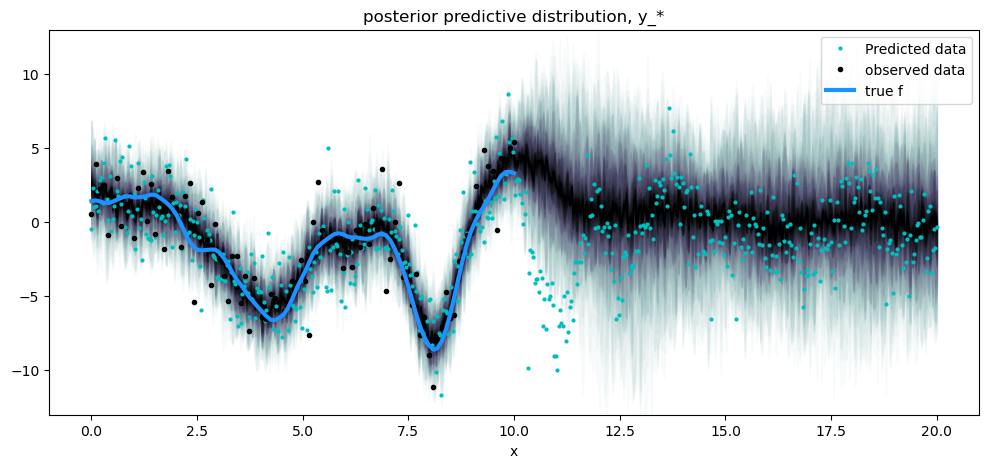
请注意,后验预测密度比无噪声函数的条件分布更宽,并反映了噪声数据的预测分布,标记为黑点。浅色点并不完全遵循预测密度的扩散,因为它们是从 GP 加噪声的后验中抽取的一个样本。
使用 .predict#
我们可以使用 .predict 方法返回给定特定 point 的均值和方差。
# predict
with model:
mu, var = gp.predict(
X_new, point=az.extract(marginal_post.posterior.sel(draw=[0])).squeeze(), diag=True
)
sd = np.sqrt(var)
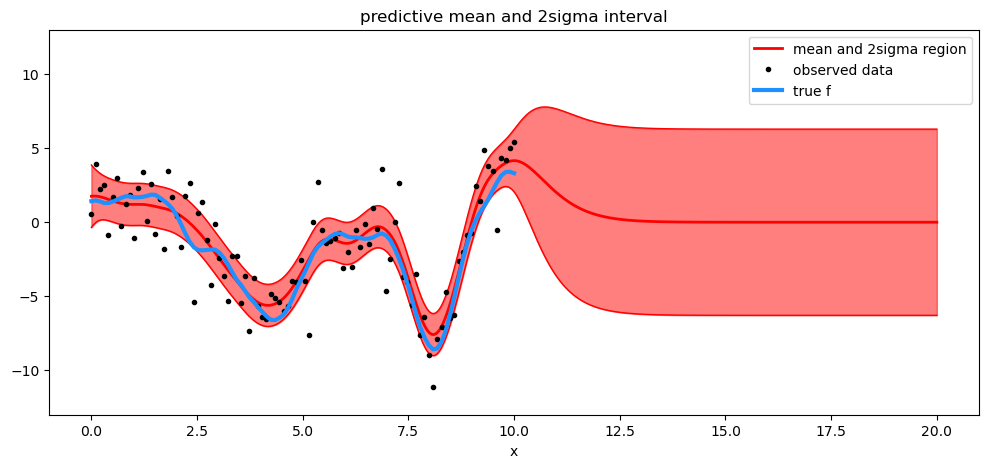
# draw plot
fig = plt.figure(figsize=(12, 5))
ax = fig.gca()
# plot mean and 2sigma intervals
plt.plot(X_new, mu, "r", lw=2, label="mean and 2σ region")
plt.plot(X_new, mu + 2 * sd, "r", lw=1)
plt.plot(X_new, mu - 2 * sd, "r", lw=1)
plt.fill_between(X_new.flatten(), mu - 2 * sd, mu + 2 * sd, color="r", alpha=0.5)
# plot original data and true function
plt.plot(X, y, "ok", ms=3, alpha=1.0, label="observed data")
plt.plot(X, f_true, "dodgerblue", lw=3, label="true f")
plt.xlabel("x")
plt.ylim([-13, 13])
plt.title("predictive mean and 2sigma interval")
plt.legend();
水印#
%load_ext watermark
%watermark -n -u -v -iv -w
Last updated: Mon Jun 05 2023
Python implementation: CPython
Python version : 3.11.3
IPython version : 8.13.2
arviz : 0.15.1
scipy : 1.10.1
pymc : 5.3.0
pandas : 2.0.2
numpy : 1.24.3
matplotlib: 3.7.1
Watermark: 2.3.1