Student-t 过程#
PyMC 也包括 T 过程先验。它们是将高斯过程先验推广到多元 Student’s T 分布。用法与 gp.Latent 相同,除了在模型中指定时,它们需要自由度参数。有关更多信息,请参阅 Rasmussen+Williams 的第 9 章和 Shah et al.。
请注意,T 过程的加法性与 GP 不同,因此不支持添加 TP 对象。
TP 先验的样本#
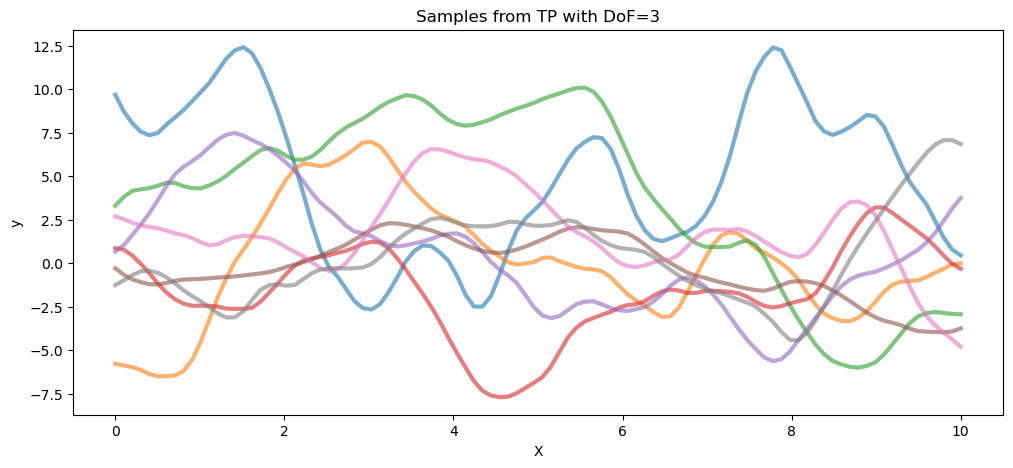
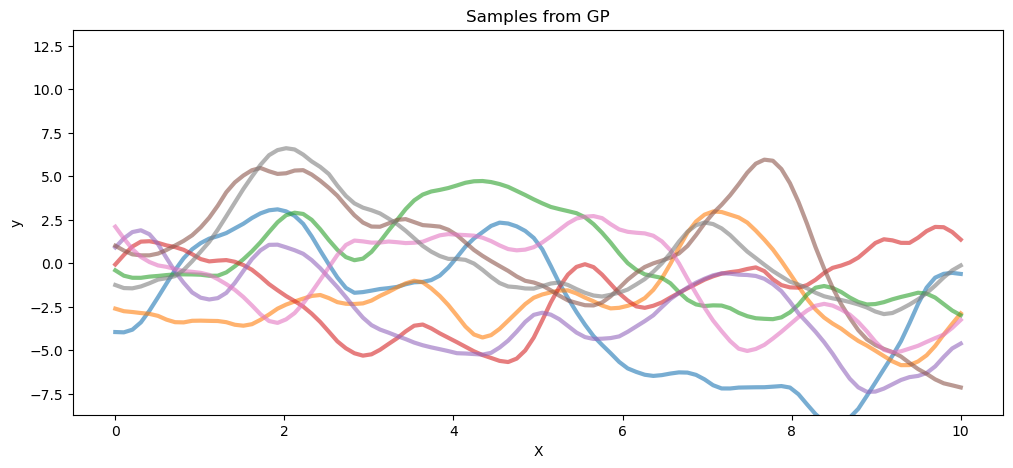
以下代码从自由度为 3 的 T 过程先验和高斯过程(两者都具有相同的协方差矩阵)中抽取样本。
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
import pymc as pm
import pytensor.tensor as pt
from pymc.gp.util import plot_gp_dist
%matplotlib inline
# set the seed
np.random.seed(1)
n = 100 # The number of data points
X = np.linspace(0, 10, n)[:, None] # The inputs to the GP, they must be arranged as a column vector
# Define the true covariance function and its parameters
ell_true = 1.0
eta_true = 3.0
cov_func = eta_true**2 * pm.gp.cov.Matern52(1, ell_true)
# A mean function that is zero everywhere
mean_func = pm.gp.mean.Zero()
# The latent function values are one sample from a multivariate normal
# Note that we have to call `eval()` because PyMC3 built on top of Theano
tp_samples = pm.draw(pm.MvStudentT.dist(mu=mean_func(X).eval(), scale=cov_func(X).eval(), nu=3), 8)
## Plot samples from TP prior
fig = plt.figure(figsize=(12, 5))
ax0 = fig.gca()
ax0.plot(X.flatten(), tp_samples.T, lw=3, alpha=0.6)
ax0.set_xlabel("X")
ax0.set_ylabel("y")
ax0.set_title("Samples from TP with DoF=3")
gp_samples = pm.draw(pm.MvNormal.dist(mu=mean_func(X).eval(), cov=cov_func(X).eval()), 8)
fig = plt.figure(figsize=(12, 5))
ax1 = fig.gca()
ax1.plot(X.flatten(), gp_samples.T, lw=3, alpha=0.6)
ax1.set_xlabel("X")
ax1.set_ylabel("y")
ax1.set_ylim(ax0.get_ylim())
ax1.set_title("Samples from GP");
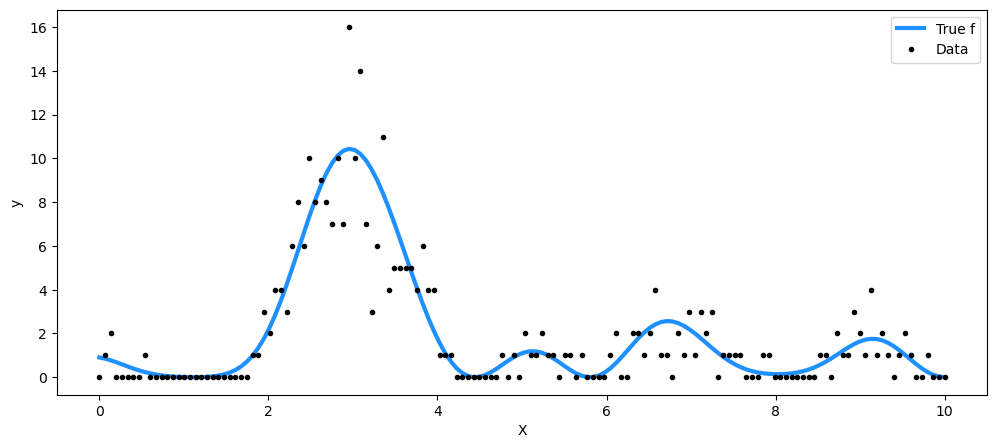
由 T 过程生成的泊松数据#
对于泊松率,我们取 T 过程先验表示的函数的平方。
np.random.seed(7)
n = 150 # The number of data points
X = np.linspace(0, 10, n)[:, None] # The inputs to the GP, they must be arranged as a column vector
# Define the true covariance function and its parameters
ell_true = 1.0
eta_true = 3.0
cov_func = eta_true**2 * pm.gp.cov.ExpQuad(1, ell_true)
# A mean function that is zero everywhere
mean_func = pm.gp.mean.Zero()
# The latent function values are one sample from a multivariate normal
# Note that we have to call `eval()` because PyMC3 built on top of Theano
f_true = pm.draw(pm.MvStudentT.dist(mu=mean_func(X).eval(), scale=cov_func(X).eval(), nu=3), 1)
y = np.random.poisson(f_true**2)
fig = plt.figure(figsize=(12, 5))
ax = fig.gca()
ax.plot(X, f_true**2, "dodgerblue", lw=3, label="True f")
ax.plot(X, y, "ok", ms=3, label="Data")
ax.set_xlabel("X")
ax.set_ylabel("y")
plt.legend();
with pm.Model() as model:
ell = pm.Gamma("ell", alpha=2, beta=2)
eta = pm.HalfCauchy("eta", beta=3)
cov = eta**2 * pm.gp.cov.ExpQuad(1, ell)
# informative prior on degrees of freedom < 5
nu = pm.Gamma("nu", alpha=2, beta=1)
tp = pm.gp.TP(scale_func=cov, nu=nu)
f = tp.prior("f", X=X)
pm.Poisson("y", mu=pt.square(f), observed=y)
tr = pm.sample(target_accept=0.9, nuts_sampler="nutpie", chains=2)
/var/home/fonnesbeck/repos/pymc-examples/.pixi/envs/default/lib/python3.12/site-packages/pytensor/graph/rewriting/basic.py:121: UserWarning: A Supervisor feature is missing from FunctionGraph(AdvancedSetSubtensor(Alloc(0.0, *2 -> Shape_i{0}(*0-<Vector(float64, shape=(?,))>), *2), *0-<Vector(float64, shape=(?,))>, *1 -> ARange{dtype='int64'}(0, *2, 1), *1)).
return self.apply(fgraph, *args, **kwargs)
/var/home/fonnesbeck/repos/pymc-examples/.pixi/envs/default/lib/python3.12/site-packages/pytensor/link/numba/dispatch/basic.py:377: UserWarning: Numba will use object mode to run AdvancedSetSubtensor's perform method
warnings.warn(
/var/home/fonnesbeck/repos/pymc-examples/.pixi/envs/default/lib/python3.12/site-packages/pytensor/graph/rewriting/basic.py:121: UserWarning: A Supervisor feature is missing from FunctionGraph(AdvancedSetSubtensor(Alloc(0.0, *1 -> Shape_i{0}(*0-<Vector(float64, shape=(?,))>), *1), *0-<Vector(float64, shape=(?,))>, *2 -> ARange{dtype='int64'}(0, *1, 1), *2)).
return self.apply(fgraph, *args, **kwargs)
/var/home/fonnesbeck/repos/pymc-examples/.pixi/envs/default/lib/python3.12/site-packages/pytensor/link/numba/dispatch/basic.py:377: UserWarning: Numba will use object mode to run AdvancedSetSubtensor's perform method
warnings.warn(
采样器进度
总链数:2
活动链数:0
已完成链数:2
采样 4 分钟
预计完成时间:now
| 进度 | 抽取 | 发散 | 步长 | 梯度/抽取 |
|---|---|---|---|---|
| 2000 | 21 | 0.07 | 127 | |
| 2000 | 13 | 0.08 | 127 |
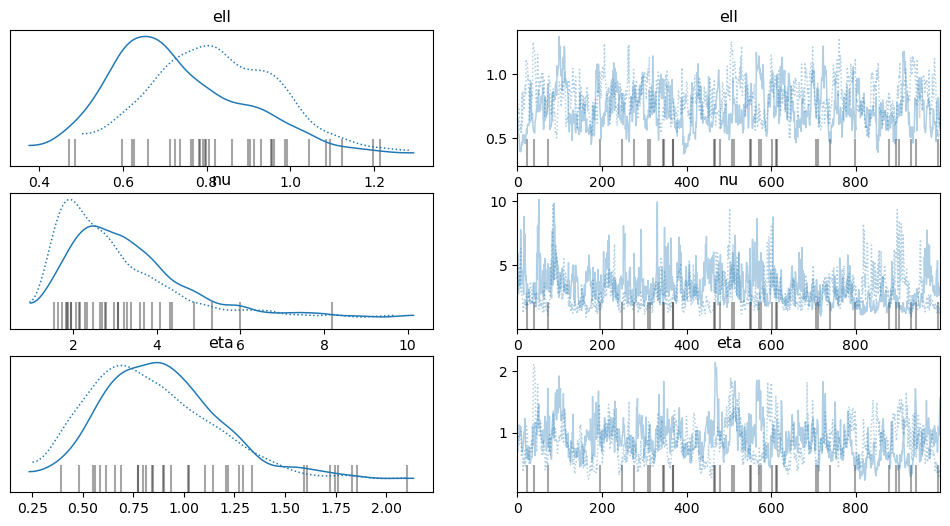
az.plot_trace(tr, var_names=["ell", "nu", "eta"]);
n_new = 200
X_new = np.linspace(0, 15, n_new)[:, None]
# add the GP conditional to the model, given the new X values
with model:
f_pred = tp.conditional("f_pred", X_new)
# Sample from the GP conditional distribution
with model:
pm.sample_posterior_predictive(tr, var_names=["f_pred"], extend_inferencedata=True)
Sampling: [f_pred]
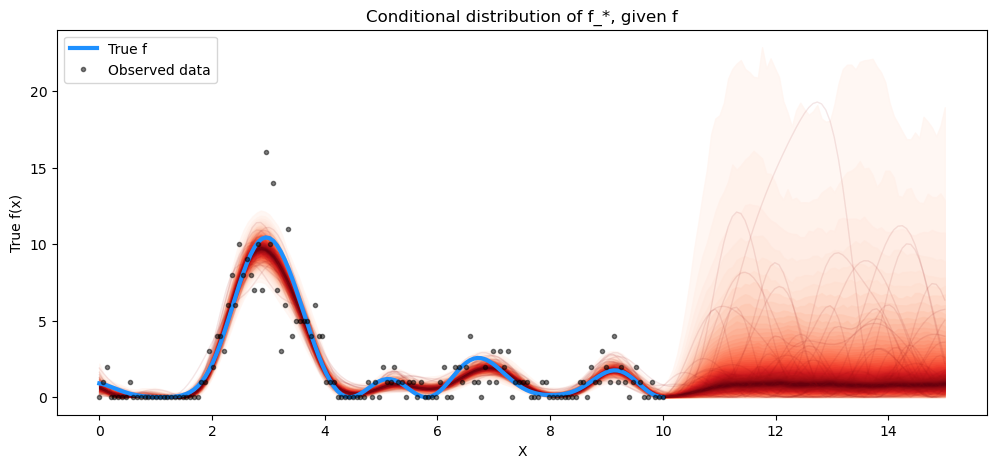
fig = plt.figure(figsize=(12, 5))
ax = fig.gca()
f_pred_samples = np.square(
az.extract(tr.posterior_predictive).astype(np.float32)["f_pred"].values
).T
plot_gp_dist(ax, f_pred_samples, X_new)
plt.plot(X, np.square(f_true), "dodgerblue", lw=3, label="True f")
plt.plot(X, y, "ok", ms=3, alpha=0.5, label="Observed data")
plt.xlabel("X")
plt.ylabel("True f(x)")
plt.title("Conditional distribution of f_*, given f")
plt.legend();
参考文献#
%load_ext watermark
%watermark -n -u -v -iv -w
Last updated: Fri Sep 20 2024
Python implementation: CPython
Python version : 3.12.5
IPython version : 8.27.0
matplotlib: 3.9.2
arviz : 0.19.0
numpy : 1.26.4
pymc : 5.16.2
pytensor : 2.25.4
Watermark: 2.4.3