更新先验#
在本笔记本中,我们将展示原则上如何在新数据可用时更新先验。
警告
此示例为 Interpolated 类提供了一个非常好的用例,我们将在下面看到。但是,在实践中这样做可能不是一个好主意,不仅因为 KDE 被用于计算后验的 pdf 值,而且主要是因为用作先验的 Interpolated 分布是单维和不相关的。因此,即使在边际上完美拟合,它们也并没有真正将我们从先前的后验中获得的所有信息纳入模型,尤其是在后验变量相关时。有关该主题的精彩讨论,请参阅 Oriol Abril 的一些维度陷阱 博文,作者是 Oriol Abril。
import arviz as az
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import pymc as pm
import pytensor.tensor as pt
from scipy import stats
from tqdm.notebook import trange
az.style.use("arviz-white")
%config InlineBackend.figure_format = "retina"
rng: np.random.Generator = np.random.default_rng(seed=42)
生成数据#
# True parameter values
alpha_true = 5
beta0_true = 7
beta1_true = 13
sigma_true = 2
# Size of dataset
size = 100
# Predictor variable
X1 = rng.normal(size=size)
X2 = rng.normal(size=size) * 0.2
# Simulate outcome variable
Y = alpha_true + beta0_true * X1 + beta1_true * X2 + rng.normal(size=size, scale=sigma_true)
模型规范#
我们对参数的初始信念信息量很大 (sigma=1) 并且与真实值略有偏差。
with pm.Model() as model:
# Priors for unknown model parameters
alpha = pm.Normal("alpha", mu=0, sigma=5)
beta0 = pm.Normal("beta0", mu=0, sigma=5)
beta1 = pm.Normal("beta1", mu=0, sigma=5)
sigma = pm.HalfNormal("sigma", sigma=1)
# Expected value of outcome
mu = alpha + beta0 * X1 + beta1 * X2
# Likelihood (sampling distribution) of observations
Y_obs = pm.Normal("Y_obs", mu=mu, sigma=sigma, observed=Y)
# draw 2_000 posterior samples
trace = pm.sample(
tune=1_500, draws=2_000, target_accept=0.9, progressbar=False, random_seed=rng
)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, beta0, beta1, sigma]
Sampling 4 chains for 1_500 tune and 2_000 draw iterations (6_000 + 8_000 draws total) took 2 seconds.
axes = az.plot_trace(
data=trace,
compact=True,
lines=[
("alpha", {}, alpha_true),
("beta0", {}, beta0_true),
("beta1", {}, beta1_true),
("sigma", {}, sigma_true),
],
backend_kwargs={"figsize": (12, 9), "layout": "constrained"},
)
plt.gcf().suptitle("Trace", fontsize=16);
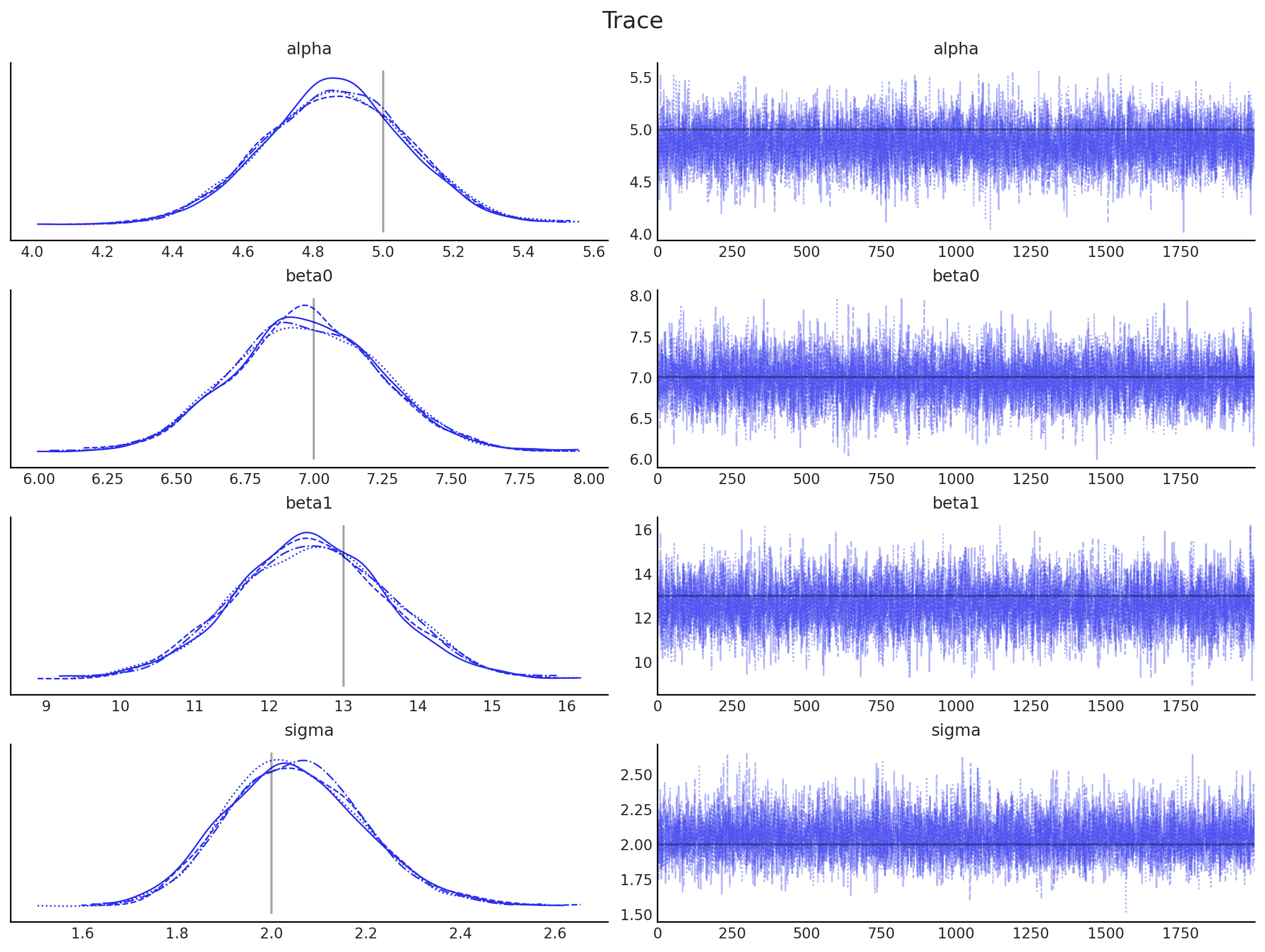
为了更新我们对参数的信念,我们使用后验分布,这将用作下一次推断的先验分布。用于每次推断迭代的数据必须独立于先前的迭代,否则相同的(可能错误的)信念会被一遍又一遍地注入到系统中,放大误差并误导推断。通过确保数据是独立的,系统应该收敛到真实的参数值。
因为我们从后验分布中抽取样本(如上图右侧所示),所以我们需要估计它们的概率密度(如上图左侧所示)。 核密度估计 (KDE) 是一种实现此目的的方法,我们将在此处使用此技术。无论如何,它是一种无法用解析方式表达的经验分布。幸运的是,PyMC 提供了一种通过 Interpolated 类使用自定义分布的方法。
def from_posterior(param, samples):
smin, smax = samples.min().item(), samples.max().item()
width = smax - smin
x = np.linspace(smin, smax, 100)
y = stats.gaussian_kde(samples)(x)
# what was never sampled should have a small probability but not 0,
# so we'll extend the domain and use linear approximation of density on it
x = np.concatenate([[x[0] - 3 * width], x, [x[-1] + 3 * width]])
y = np.concatenate([[0], y, [0]])
return pm.Interpolated(param, x, y)
现在我们只需要生成更多数据并构建我们的贝叶斯模型,以便当前迭代的先验分布是来自先前迭代的后验分布。仍然可以继续使用 NUTS 抽样方法,因为 Interpolated 类实现了 Hamiltonian Monte Carlo 采样器所需的梯度计算。
traces = [trace]
n_iterations = 10
for _ in trange(n_iterations):
# generate more data
X1 = rng.normal(size=size)
X2 = rng.normal(size=size) * 0.2
Y = alpha_true + beta0_true * X1 + beta1_true * X2 + rng.normal(size=size, scale=sigma_true)
with pm.Model() as model:
# Priors are posteriors from previous iteration
alpha = from_posterior("alpha", az.extract(trace, group="posterior", var_names=["alpha"]))
beta0 = from_posterior("beta0", az.extract(trace, group="posterior", var_names=["beta0"]))
beta1 = from_posterior("beta1", az.extract(trace, group="posterior", var_names=["beta1"]))
sigma = from_posterior("sigma", az.extract(trace, group="posterior", var_names=["sigma"]))
# Expected value of outcome
mu = alpha + beta0 * X1 + beta1 * X2
# Likelihood (sampling distribution) of observations
Y_obs = pm.Normal("Y_obs", mu=mu, sigma=sigma, observed=Y)
# draw 2_000 posterior samples
trace = pm.sample(
tune=1_500, draws=2_000, target_accept=0.9, progressbar=False, random_seed=rng
)
traces.append(trace)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, beta0, beta1, sigma]
Sampling 4 chains for 1_500 tune and 2_000 draw iterations (6_000 + 8_000 draws total) took 2 seconds.
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, beta0, beta1, sigma]
Sampling 4 chains for 1_500 tune and 2_000 draw iterations (6_000 + 8_000 draws total) took 2 seconds.
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, beta0, beta1, sigma]
Sampling 4 chains for 1_500 tune and 2_000 draw iterations (6_000 + 8_000 draws total) took 2 seconds.
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, beta0, beta1, sigma]
Sampling 4 chains for 1_500 tune and 2_000 draw iterations (6_000 + 8_000 draws total) took 2 seconds.
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, beta0, beta1, sigma]
Sampling 4 chains for 1_500 tune and 2_000 draw iterations (6_000 + 8_000 draws total) took 2 seconds.
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, beta0, beta1, sigma]
Sampling 4 chains for 1_500 tune and 2_000 draw iterations (6_000 + 8_000 draws total) took 2 seconds.
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, beta0, beta1, sigma]
Sampling 4 chains for 1_500 tune and 2_000 draw iterations (6_000 + 8_000 draws total) took 4 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for details
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, beta0, beta1, sigma]
Sampling 4 chains for 1_500 tune and 2_000 draw iterations (6_000 + 8_000 draws total) took 3 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for details
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, beta0, beta1, sigma]
Sampling 4 chains for 1_500 tune and 2_000 draw iterations (6_000 + 8_000 draws total) took 3 seconds.
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, beta0, beta1, sigma]
Sampling 4 chains for 1_500 tune and 2_000 draw iterations (6_000 + 8_000 draws total) took 5 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for details
fig, ax = plt.subplots(nrows=4, ncols=1, figsize=(12, 12), sharex=False, sharey=False)
cmap = mpl.cm.viridis
for i, (param, true_value) in enumerate(
zip(["alpha", "beta0", "beta1", "sigma"], [alpha_true, beta0_true, beta1_true, sigma_true])
):
for update_i, trace in enumerate(traces):
samples = az.extract(trace, group="posterior", var_names=param)
smin, smax = np.min(samples), np.max(samples)
x = np.linspace(smin, smax, 100)
y = stats.gaussian_kde(samples)(x)
ax[i].plot(x, y, color=cmap(1 - update_i / len(traces)))
ax[i].axvline(true_value, c="k")
ax[i].set(title=param)

您可以重新执行最后两个单元格以生成更多更新。
有趣的是,我们参数的后验分布趋于以它们的真实值(垂直线)为中心,并且分布变得越来越细。这意味着我们每次都变得更加自信,并且我们最初拥有的(错误)信念被我们纳入的新数据冲刷掉。
并非万能药
请注意,尽管迭代看起来似乎有所改进,但其中一些看起来不太好,甚至有时看起来会倒退。除了笔记本开头指出的原因外,过程中还有几个关键步骤涉及随机性。因此,应该预期事情平均而言会有所改善。
新的观察是随机的。如果在初始迭代中,我们获得的值更接近分布的主体,然后我们连续从正尾获得几个值,那么我们累积了几个来自尾部的抽样的迭代可能会有偏差,并且“看起来比以前的迭代更差”。
MCMC 是随机的。即使它收敛,MCMC 也是一个随机过程,因此对
pymc.sample的不同调用将返回以精确后验为中心但并非总是相同的值;应该预期的变化有多大可以使用arviz.mcse()检查。KDE 也将这种通常可以忽略不计但又存在的后验估计不确定性纳入其中,因此生成的 Interpolated 分布也会如此。
替代方法
在 pymc-extras 中还有另一种替代方法,通过函数 prior_from_idata() 可以实现类似的功能。此函数
使用 MvNormal 近似从后验创建先验。该近似使用 MvNormal 分布。请记住,此函数仅适用于单峰后验,并且在发生复杂的交互时会失败。此外,如果检索到的变量受到约束,您应该为该变量指定一个变换,例如标准差后验的
log()。
许可声明#
此示例库中的所有笔记本均根据 MIT 许可证 提供,该许可证允许修改和再分发用于任何用途,前提是保留版权和许可声明。
引用 PyMC 示例#
要引用此笔记本,请使用 Zenodo 为 pymc-examples 存储库提供的 DOI。
重要提示
许多笔记本都改编自其他来源:博客、书籍…… 在这种情况下,您也应该引用原始来源。
另请记住引用您的代码使用的相关库。
这是一个 bibtex 中的引用模板
@incollection{citekey,
author = "<notebook authors, see above>",
title = "<notebook title>",
editor = "PyMC Team",
booktitle = "PyMC examples",
doi = "10.5281/zenodo.5654871"
}
渲染后可能看起来像