孟加拉国生育率再探#
utils.draw_causal_graph(
edge_list=[
("A", "C"),
("K", "C"),
("U", "C"),
("D", "C"),
("D", "U"),
("U", "K"),
("A", "K"),
("D", "K"),
]
)
估计量 1:每个地区 \(D\) 的避孕药具使用率 \(C\)
估计量 2:“城市化”的影响 \(U\)
估计量 3:孩子数量 \(K\) 和年龄 \(A\) 的影响
地区是聚类#
农村/城市细分#
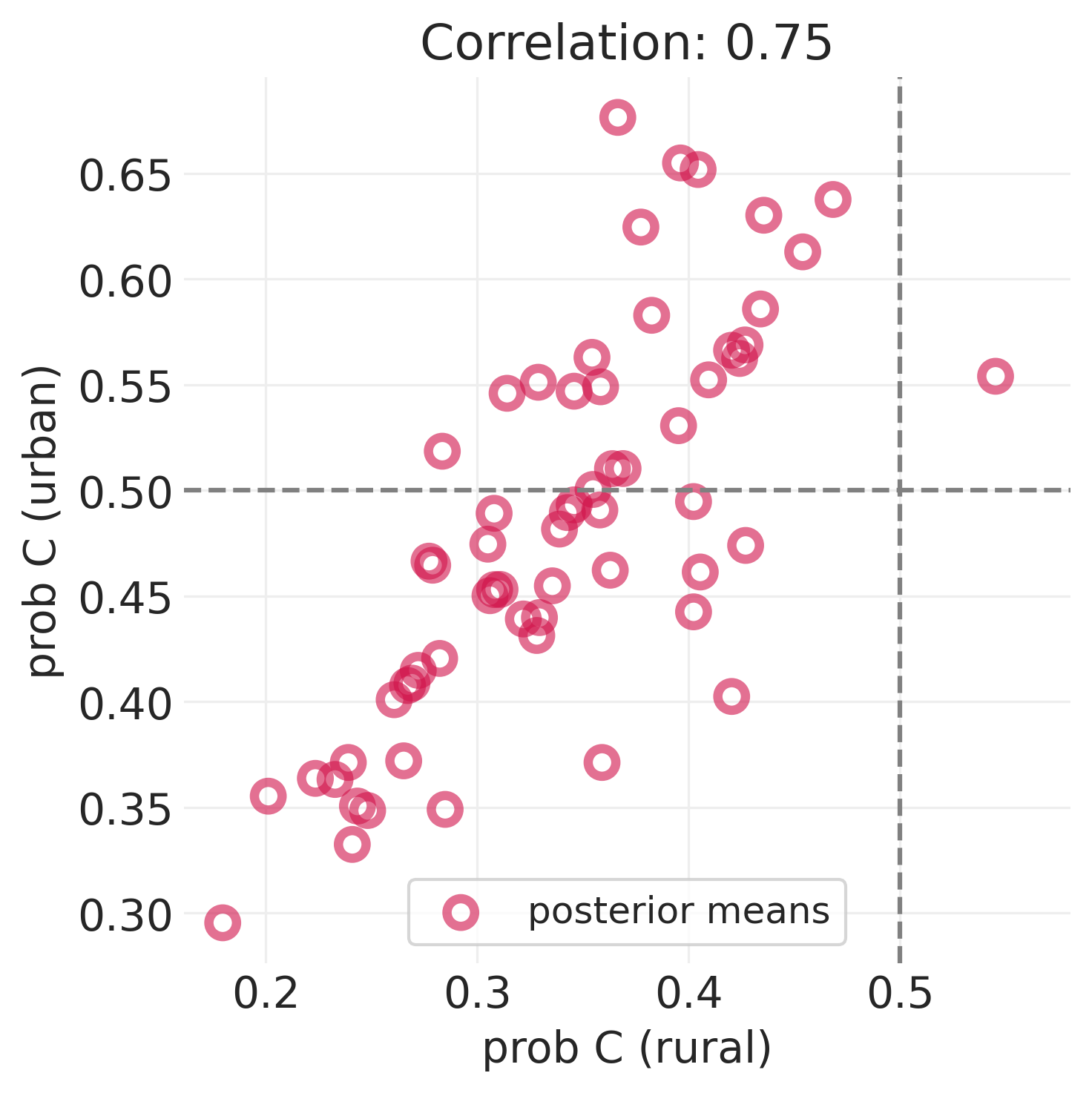
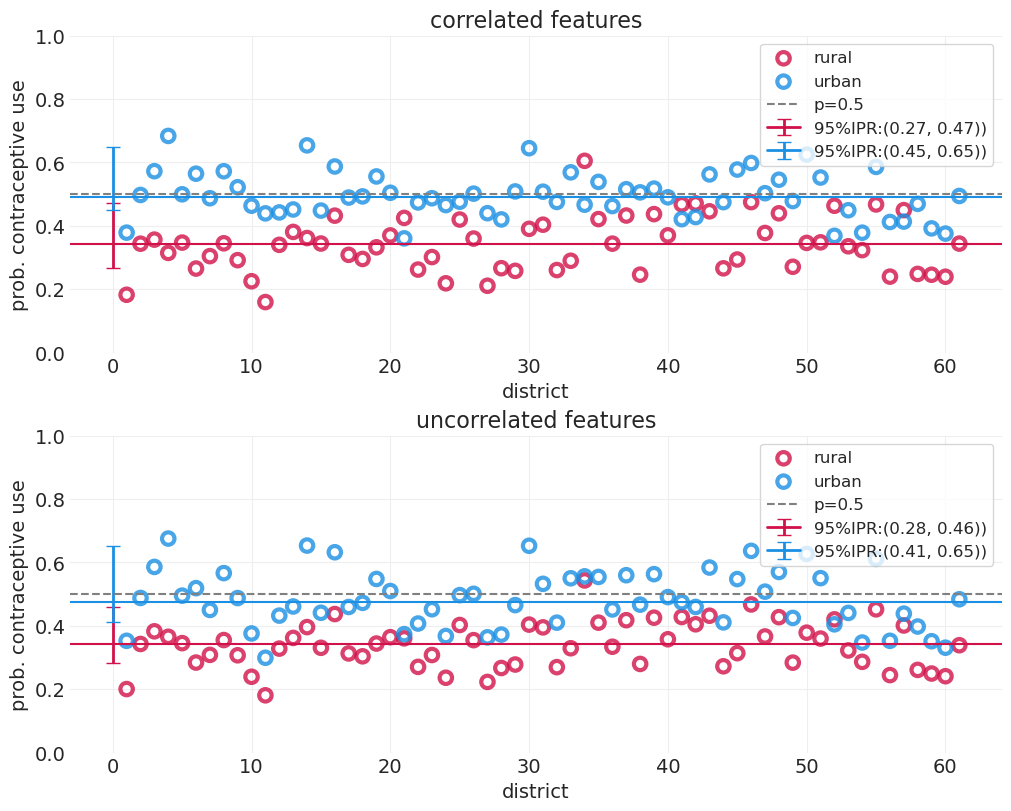
下面我们展示了农村地区(仅包括地区级截距 \(\alpha_{D[i]}\))与城市地区(包括城市地区的额外截距 \(\beta_{D[i]}\))组的避孕药具使用率。这些图是在 第 13 讲 - 多层冒险 中生成的。
utils.display_image("fertility_posterior_means_rural_urban.png", width=1000)
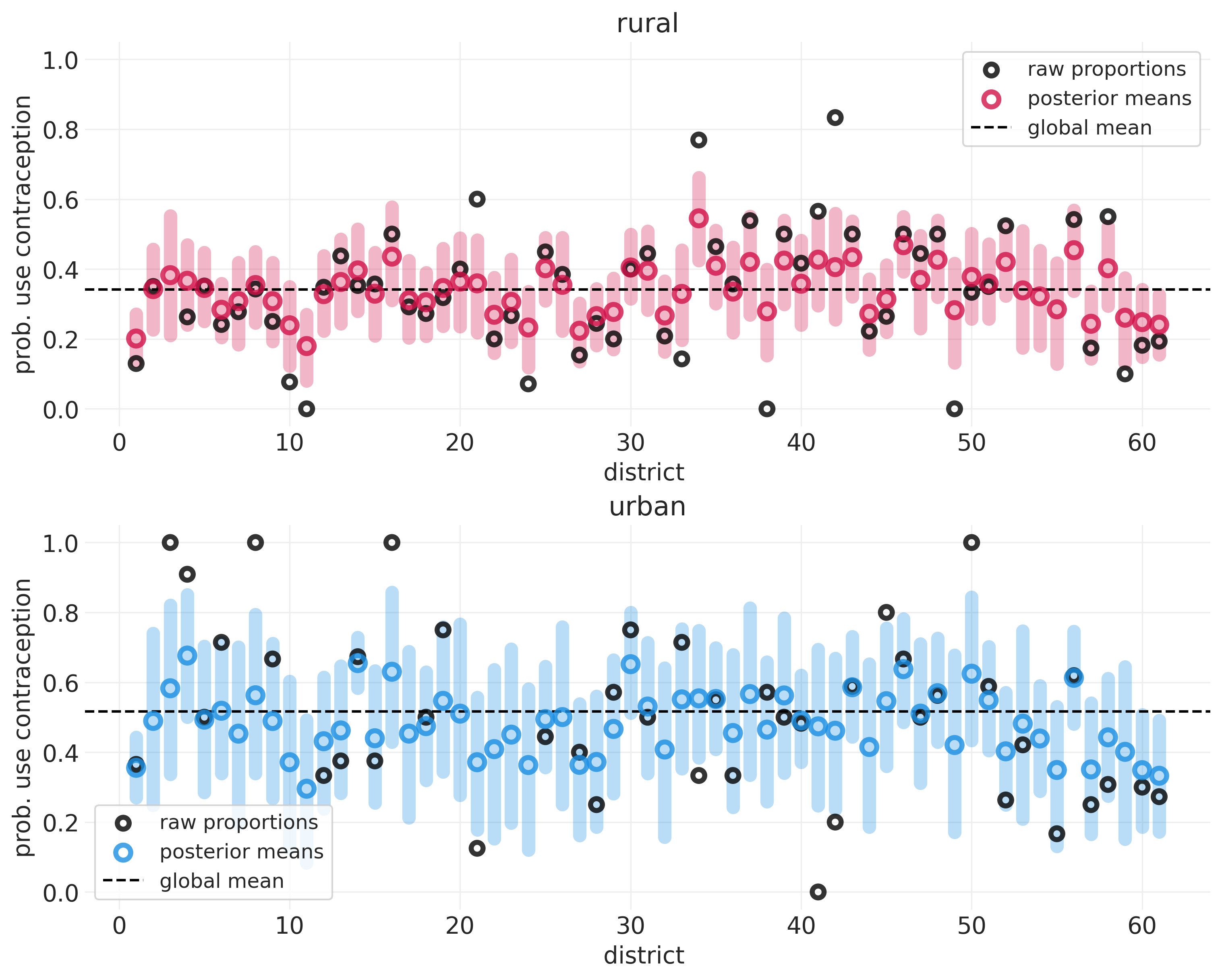
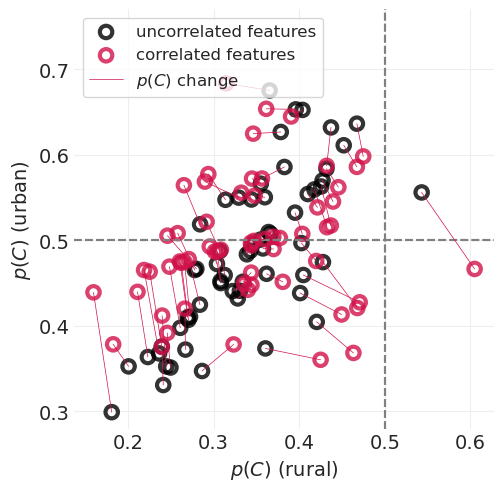
该图显示
平均而言,农村人口比城市地区人口更不可能使用避孕措施
顶部图中的虚线低于底部图中的虚线
城市样本较少,因此城市地区的不确定性范围较大
与农村地区相比,与城市人口相关的误差蓝色条更大,农村地区采样的民意调查更多。
🤔#
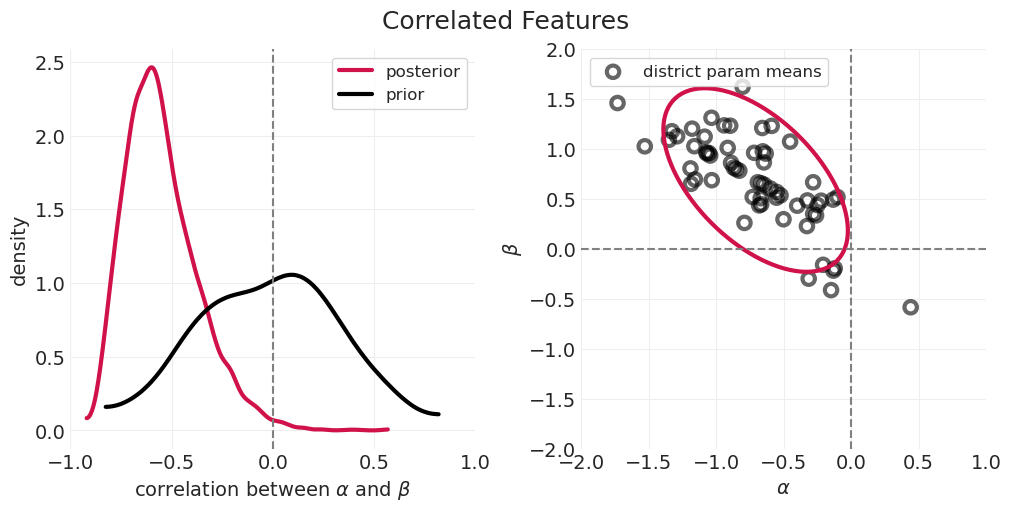
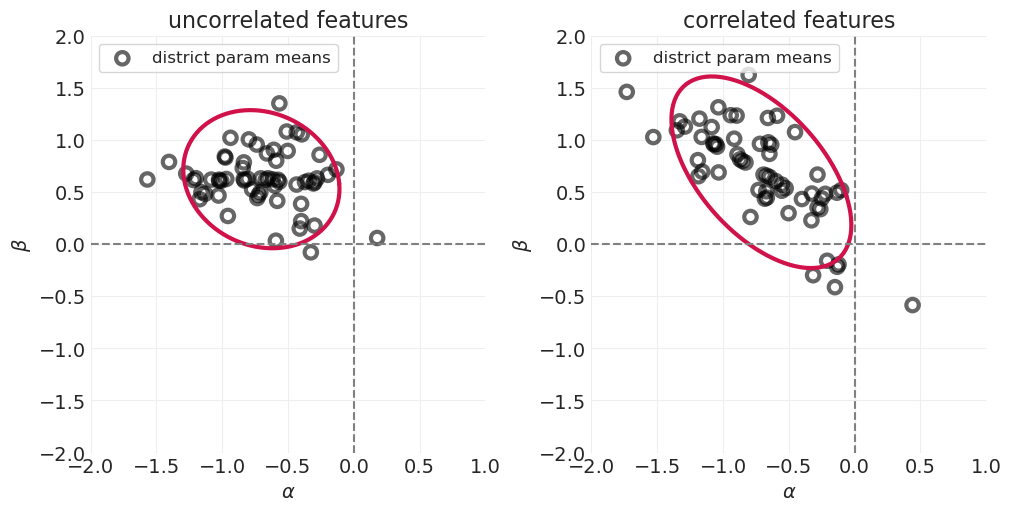
如果我们知道农村地区的避孕药具使用率,由于这种相关性,您可以更好地猜测城市地区的避孕药具使用率
我们没有让 Golem(即我们的统计模型)利用这种特征相关性,因此是在浪费信息
本讲座的重点是构建可以捕获特征之间相关信息的统计模型。
通过跨特征的部分合并,使推断更有效
估计相关性和部分合并演示
McElreath 继续展示了贝叶斯更新在具有相关特征的模型中的演示。它非常棒,我建议多次回顾该演示,但我太懒了,无法实现它(也没有聪明到在没有动画的情况下做到这一点,我正在尝试避免动画)。它将添加到 TODO 列表中。
奖励:非居中(又名变换)先验#
不方便的后验#
由负对数似然中的陡峭曲率引起的不高效 MCMC
哈密顿蒙特卡洛在探索陡峭表面时遇到困难
导致“发散”跃迁
“冲破滑板公园的墙壁”
当哈密顿量在提议的开始/结束之间发生剧烈变化时检测到
变换先验以使其成为“更平滑的滑板公园”有所帮助
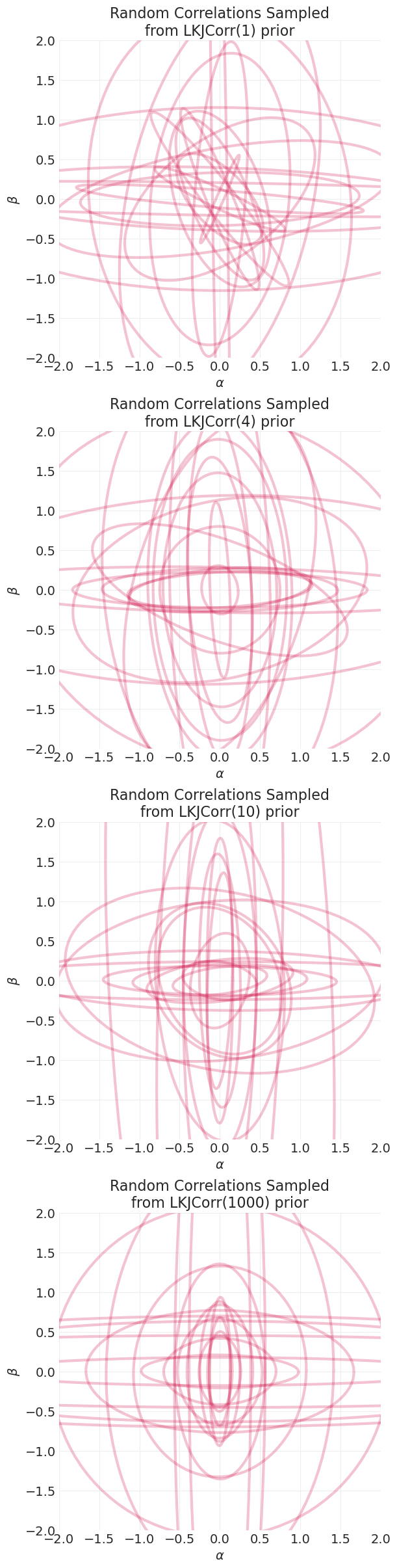
示例:魔鬼漏斗先验#
随着 \(\sigma_v\) 的增加,先验概率表面中会形成一个讨厌的槽,哈密顿动力学很难对其进行采样——即,滑板公园太陡峭和狭窄。
我太懒了,无法编写 McElreath 用来可视化每个发散路径的精美 HMC 动画。但是,我们仍然可以验证,当我们在魔鬼漏斗先验中增加 v 先验的 \(\sigma\) 时,发散的数量会增加。
居中先验模型#
from functools import partial
xs = vs = np.linspace(-4, 4, 100)
def prior_devils_funnel(xs, vs, sigma_v=0.5):
log_prior_v = stats.norm(0, sigma_v).logpdf(vs)
log_prior_x = stats.norm(0, np.exp(vs)).logpdf(xs)
log_prior = log_prior_v + log_prior_x
return np.exp(log_prior - log_prior.max())
# Loop over sigma_v, and show that divergences increase
# as the depth and narrowness of the trough increases
# for low values of v
sigma_vs = [0.5, 1, 3]
n_sigma_vs = len(sigma_vs)
fig, axs = plt.subplots(3, 1, figsize=(4, 4 * n_sigma_vs))
for ii, sigma_v in enumerate(sigma_vs):
with pm.Model() as centered_devils_funnel:
v = pm.Normal("v", 0, sigma_v)
x = pm.Normal("x", 0, pm.math.exp(v))
centered_devils_funnel_inference = pm.sample()
n_divergences = centered_devils_funnel_inference.sample_stats.diverging.sum().values
prior = partial(prior_devils_funnel, sigma_v=sigma_v)
plt.sca(axs[ii])
utils.plot_2d_function(xs, ys, prior, colors="gray", ax=axs[ii])
plt.xlabel("x")
plt.ylabel("v")
plt.title(f"$v \sim Normal(0, {sigma_v})$\n# Divergences: {n_divergences}")
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [v, x]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 1 seconds.
There were 9 divergences after tuning. Increase `target_accept` or reparameterize.
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [v, x]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 1 seconds.
There were 405 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for details
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [v, x]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 1 seconds.
There were 622 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for details
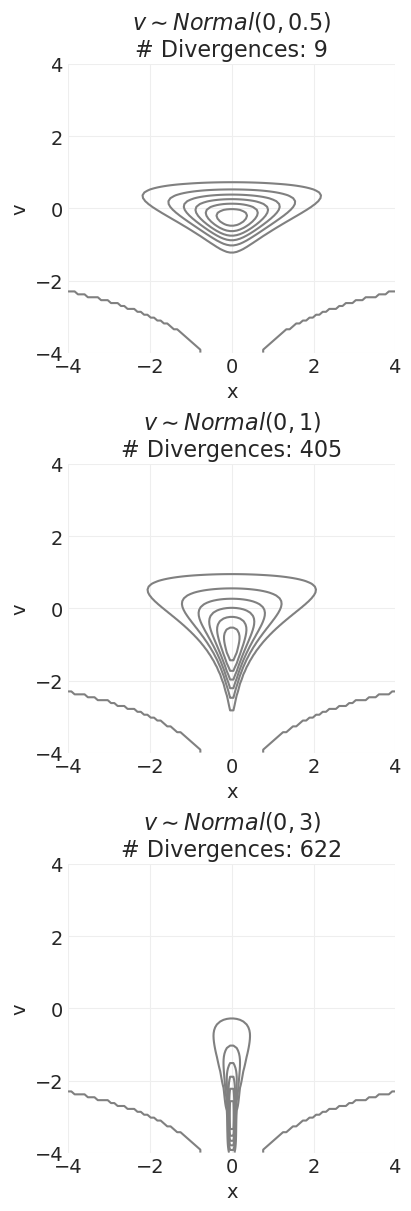
怎么办?#
更小的步长:更好地处理陡峭度,但需要更长的时间来探索后验
重新参数化(变换)以使表面更平滑
非居中先验#
我们添加一个辅助变量 \(z\),它具有平滑的概率表面。然后我们对该辅助变量进行采样,并对其进行变换以获得目标变量分布。对于魔鬼漏斗先验的情况
sigma_vs = [0.5, 1, 3]
n_sigma_vs = len(sigma_vs)
fig, axs = plt.subplots(3, 1, figsize=(4, 4 * n_sigma_vs))
def prior_noncentered_devils_funnel(xs, vs, sigma_v=0.5):
"""Reparamterize into a smoother auxilary variable space"""
log_prior_v = stats.norm(0, sigma_v).logpdf(vs)
log_prior_z = stats.norm(0, 1).logpdf(xs)
log_prior = log_prior_v + log_prior_z
return np.exp(log_prior - log_prior.max())
for ii, sigma_v in enumerate(sigma_vs):
with pm.Model() as noncentered_devils_funnel:
v = pm.Normal("v", 0, sigma_v)
z = pm.Normal("z", 0, 1)
# Record x for reporting
x = pm.Deterministic("x", z * pm.math.exp(v))
noncentered_devils_funnel_inference = pm.sample()
n_divergences = noncentered_devils_funnel_inference.sample_stats.diverging.sum().values
prior = partial(prior_noncentered_devils_funnel, sigma_v=sigma_v)
plt.sca(axs[ii])
utils.plot_2d_function(xs, ys, prior, colors="gray", ax=axs[ii])
plt.xlabel("z")
plt.ylabel("v")
plt.title(f"$v \sim$ Normal(0, {sigma_v})\n# Divergences: {n_divergences}")
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [v, z]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 1 seconds.
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [v, z]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 1 seconds.
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [v, z]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 1 seconds.
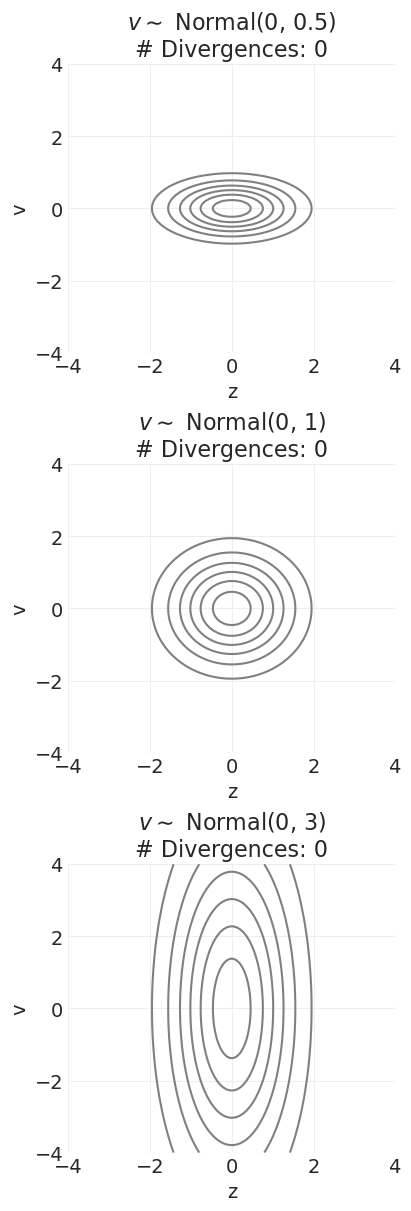
通过重新参数化,我们可以对多维正态分布进行采样,这些分布在对数空间中是更平滑的抛物线。
我们可以看到,对于所有先验方差值,发散的数量都持续减少。
检查 HMC 诊断#
az.summary(noncentered_devils_funnel_inference)["r_hat"]
v 1.0
x 1.0
z 1.0
Name: r_hat, dtype: float64
az.plot_trace(noncentered_devils_funnel_inference);
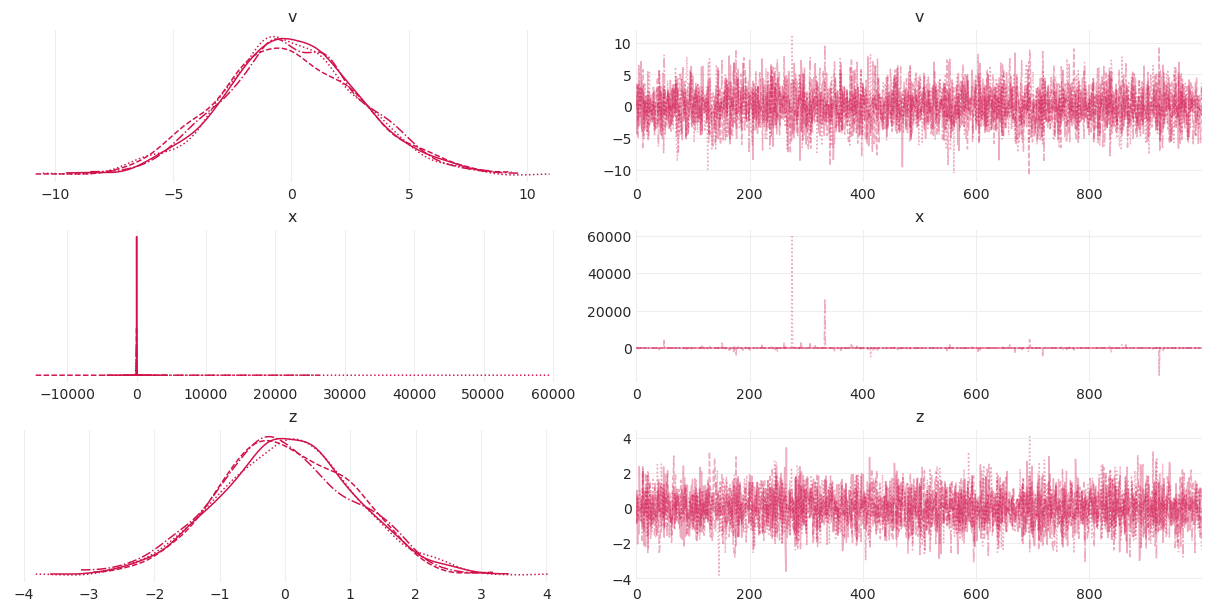
诊断看起来不错 👍
Rhats = 1\(v\) 和 \(z\) 链表现出“模糊毛毛虫”行为
Cholesky 因子和相关矩阵#
Cholesky 因子 \(\bf L\) 提供了一种有效的方法来编码相关矩阵 \(\Omega\)(比完整的相关矩阵需要更少的浮点数)
\(\Omega = \bf LL^T\)
\(\bf L\) 是一个下三角矩阵
我们可以使用 \(\Omega\) 的 Cholesky 分解 \(\bf L\) 对具有相关性 \(\Omega\) 和标准差 \(\sigma_i\) 的数据进行采样,如下所示
其中 \(\bf Z\) 是从标准正态分布中采样的 z 分数矩阵
演示从 Cholesky 因子重建相关矩阵#
orig_corr = np.array([[1, 0.6], [0.6, 1]])
print("Ground-truth Correlation Matrix\n", orig_corr)
L = np.linalg.cholesky(orig_corr)
print("\nCholesky Matrix of correlation matrix, L\n", L)
print("\nReconstructed correlation matrix from L\n", L @ L.T)
Ground-truth Correlation Matrix
[[1. 0.6]
[0.6 1. ]]
Cholesky Matrix of correlation matrix, L
[[1. 0. ]
[0.6 0.8]]
Reconstructed correlation matrix from L
[[1. 0.6]
[0.6 1. ]]
演示从 z 分数和 Cholesky 因子采样随机相关矩阵#
np.random.seed(12345)
N = 100_000
orig_sigmas = [2, 0.5]
# Matrix of z-score samples
Z = stats.norm().rvs(size=(2, N))
print("Raw z-score samples are indpendent\n", np.corrcoef(Z[0], Z[1]))
# Transform Z-scores using the Cholesky factorization of the correlation matrix
sLZ = np.diag(orig_sigmas) @ L @ Z
corr_sLZ = np.corrcoef(sLZ[0], sLZ[1])
print("\nCholesky-transformed z-scores have original correlation encoded\n", corr_sLZ)
assert np.isclose(orig_corr[0, 1], corr_sLZ[0, 1], atol=0.01)
std_sLZ = sLZ.std(axis=1)
print("\nOriginal std devs also encoded in transformed z-scores:\n", std_sLZ)
assert np.isclose(orig_sigmas[0], std_sLZ[0], atol=0.01)
assert np.isclose(orig_sigmas[1], std_sLZ[1], atol=0.01)
Raw z-score samples are indpendent
[[1. 0.00293945]
[0.00293945 1. ]]
Cholesky-transformed z-scores have original correlation encoded
[[1. 0.60268456]
[0.60268456 1. ]]
Original std devs also encoded in transformed z-scores:
[2.00212936 0.50024976]
何时使用变换先验#
取决于上下文
居中
每个聚类/子组中的大量数据
似然主导场景
非居中
许多聚类/子组,其中一些聚类中的数据稀疏
先验主导场景
许可证声明#
此示例库中的所有笔记本均根据 MIT 许可证 提供,该许可证允许修改和再分发用于任何用途,前提是保留版权和许可证声明。
引用 PyMC 示例#
要引用此笔记本,请使用 Zenodo 为 pymc-examples 存储库提供的 DOI。
重要提示
许多笔记本都改编自其他来源:博客、书籍……在这种情况下,您也应该引用原始来源。
另请记住引用您的代码使用的相关库。
这是一个 bibtex 中的引用模板
@incollection{citekey,
author = "<notebook authors, see above>",
title = "<notebook title>",
editor = "PyMC Team",
booktitle = "PyMC examples",
doi = "10.5281/zenodo.5654871"
}
一旦呈现,它可能看起来像