PyMC变分推断入门#
计算贝叶斯模型后验量的最常用策略是通过采样,特别是马尔可夫链蒙特卡洛 (MCMC) 算法。虽然采样算法和相关的计算在性能和效率上不断提高,但 MCMC 方法在数据量较大时仍然表现不佳,并且对于超过几千个观测值的情况变得难以处理。变分推断 (VI) 是一种比采样更具可扩展性的替代方案,它将计算后验分布的问题重新定义为一个优化问题。
在 PyMC 中,变分推断 API 专注于通过一套现代算法来近似后验分布。此模块的常见用例包括
从模型后验采样并计算任意表达式
进行期望、方差和其他统计量的蒙特卡洛近似
移除对 PyMC 随机节点的符号依赖性并评估表达式(使用
eval)提供连接到任意 PyTensor 代码的桥梁
%matplotlib inline
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
import pymc as pm
import pytensor
import seaborn as sns
np.random.seed(42)
分布近似#
统计学中有几种方法使用更简单的分布来近似更复杂的分布。也许最著名的例子是拉普拉斯(正态)近似。这涉及到构建目标后验的泰勒级数,但只保留二次项,并使用这些项来构建多元正态近似。
类似地,变分推断是另一种分布近似方法,其中,不是利用泰勒级数,而是选择一类近似分布,并优化其参数,使得到的分布尽可能接近后验。本质上,VI 是一种确定性近似,它对感兴趣的密度设置边界,然后使用优化从该有界集合中进行选择。
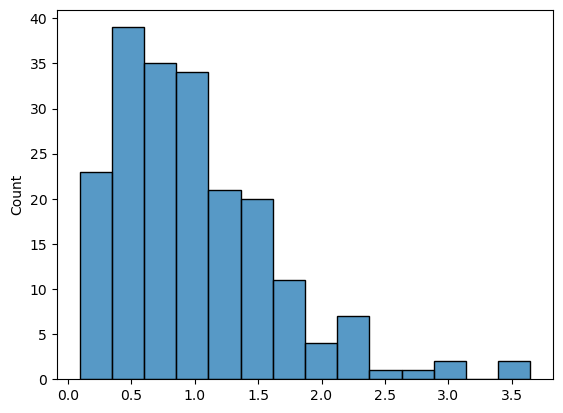
gamma_data = np.random.gamma(2, 0.5, size=200)
sns.histplot(gamma_data);
with pm.Model() as gamma_model:
alpha = pm.Exponential("alpha", 0.1)
beta = pm.Exponential("beta", 0.1)
y = pm.Gamma("y", alpha, beta, observed=gamma_data)
with gamma_model:
# mean_field = pm.fit()
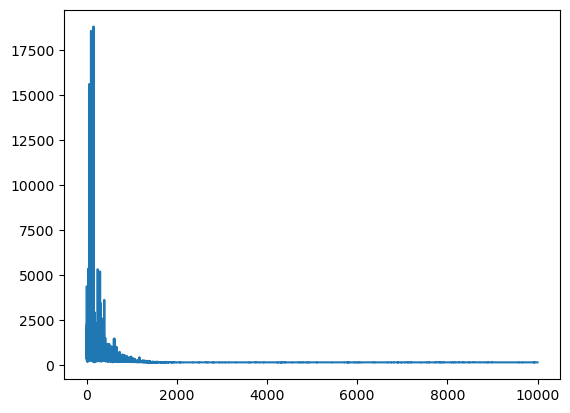
mean_field = pm.fit(obj_optimizer=pm.adagrad_window(learning_rate=1e-2))
Finished [100%]: Average Loss = 169.87
with gamma_model:
trace = pm.sample()
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, beta]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 3 seconds.
mean_field
<pymc.variational.approximations.MeanField at 0x7fca20419e50>
plt.plot(mean_field.hist);
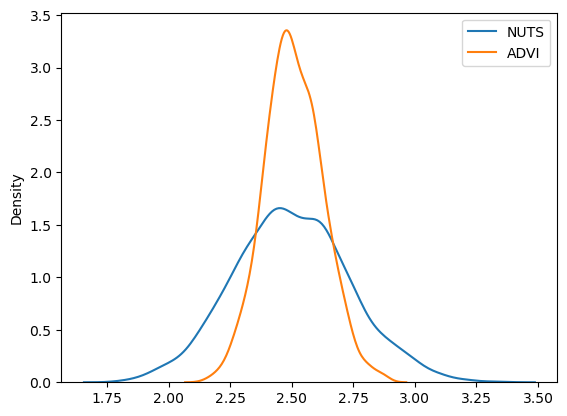
approx_sample = mean_field.sample(1000)
sns.kdeplot(trace.posterior["alpha"].values.flatten(), label="NUTS")
sns.kdeplot(approx_sample.posterior["alpha"].values.flatten(), label="ADVI")
plt.legend();
基本设置#
我们不需要复杂的模型来玩转 VI API;让我们从一个简单的混合模型开始
w = np.array([0.2, 0.8])
mu = np.array([-0.3, 0.5])
sd = np.array([0.1, 0.1])
with pm.Model() as model:
x = pm.NormalMixture("x", w=w, mu=mu, sigma=sd)
x2 = x**2
sin_x = pm.math.sin(x)
我们无法为此模型计算解析期望。但是,我们可以使用马尔可夫链蒙特卡洛方法获得近似值;让我们首先使用 NUTS。
为了允许保存表达式的样本,我们需要将它们包装在 Deterministic 对象中
with model:
pm.Deterministic("x2", x2)
pm.Deterministic("sin_x", sin_x)
with model:
trace = pm.sample(5000)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [x]
Sampling 4 chains for 1_000 tune and 5_000 draw iterations (4_000 + 20_000 draws total) took 5 seconds.
az.plot_trace(trace);
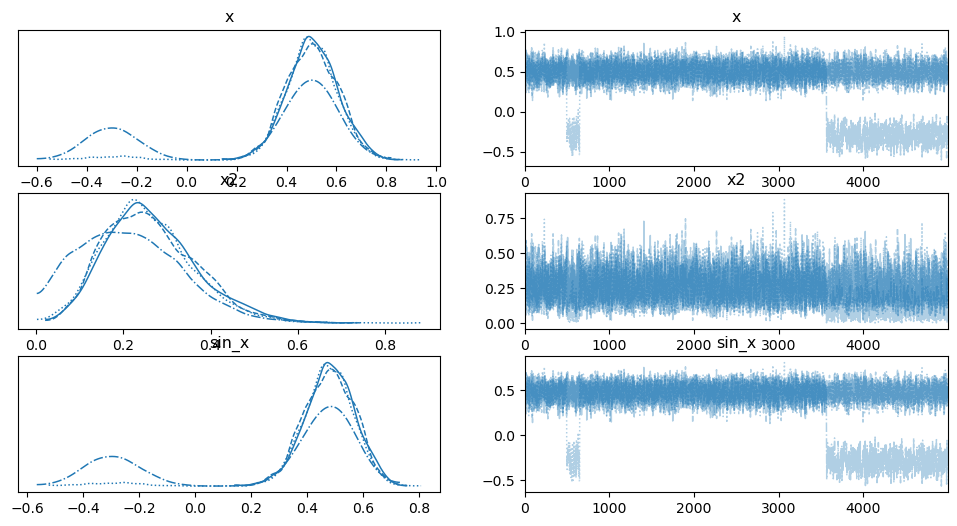
上面是 \(x^2\) 和 \(sin(x)\) 的迹。我们可以看到这个模型中存在明显的多模态。一个缺点是,您需要预先知道您想在迹中看到什么,并用 Deterministic 包裹它。
VI API 采用另一种方法:您从模型获得推断,然后在之后基于此模型计算表达式。
让我们使用相同的模型
with pm.Model() as model:
x = pm.NormalMixture("x", w=w, mu=mu, sigma=sd)
x2 = x**2
sin_x = pm.math.sin(x)
这里我们将使用自动微分变分推断 (ADVI)。
with model:
mean_field = pm.fit(method="advi")
Finished [100%]: Average Loss = 2.216
az.plot_posterior(mean_field.sample(1000), color="LightSeaGreen");
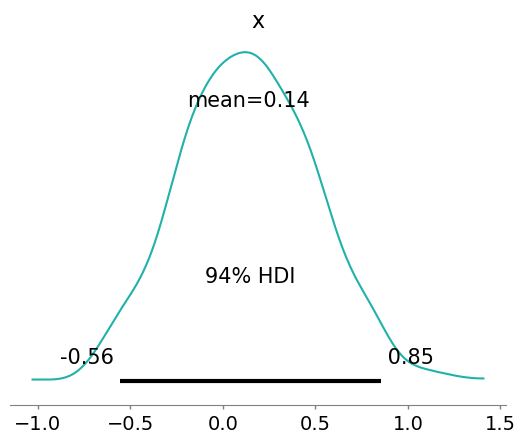
请注意,ADVI 未能近似多模态分布,因为它使用了具有单峰的高斯分布。
检查收敛#
让我们使用 CheckParametersConvergence 的默认参数,因为它们看起来是合理的。
from pymc.variational.callbacks import CheckParametersConvergence
with model:
mean_field = pm.fit(method="advi", callbacks=[CheckParametersConvergence()])
Finished [100%]: Average Loss = 2.239
我们可以通过 .hist 属性访问推断历史记录。
plt.plot(mean_field.hist);
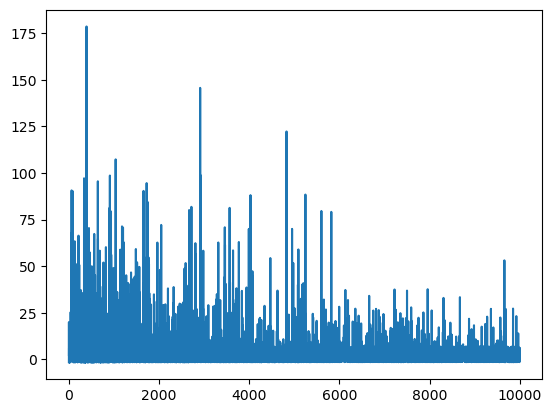
这不是一个好的收敛图,尽管我们运行了很多迭代。原因是 ADVI 近似的均值接近于零,因此采用相对差异(默认方法)对于检查收敛是不稳定的。
with model:
mean_field = pm.fit(
method="advi", callbacks=[pm.callbacks.CheckParametersConvergence(diff="absolute")]
)
Convergence achieved at 6200
Interrupted at 6,199 [61%]: Average Loss = 4.3808
plt.plot(mean_field.hist);
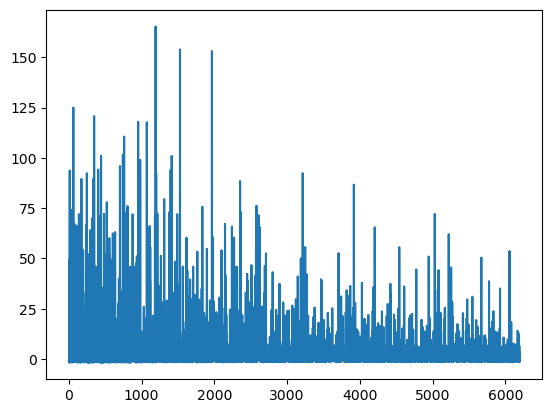
这样好多了!我们在不到 5000 次迭代后达到了收敛。
跟踪参数#
另一个有用的回调允许用户跟踪参数。它允许在推断期间跟踪任意统计量,尽管它可能很消耗内存。使用 fit 函数,我们在推断之前无法直接访问近似值。但是,跟踪参数需要访问近似值。我们可以通过使用面向对象 (OO) 的推断 API 来绕过此约束。
with model:
advi = pm.ADVI()
advi.approx
<pymc.variational.approximations.MeanField at 0x7fca1daee6a0>
不同的近似有不同的超参数。在平均场 ADVI 中,我们有 \(\rho\) 和 \(\mu\) (灵感来自 Bayes by BackProp)。
advi.approx.shared_params
{'mu': mu, 'rho': rho}
有与近似相关的相关统计量的便捷快捷方式。例如,在为 NUTS 采样指定质量矩阵时,这可能很有用
advi.approx.mean.eval(), advi.approx.std.eval()
(array([0.34]), array([0.69314718]))
我们可以将这些统计量滚动到 Tracker 回调中。
tracker = pm.callbacks.Tracker(
mean=advi.approx.mean.eval, # callable that returns mean
std=advi.approx.std.eval, # callable that returns std
)
现在,调用 advi.fit 将记录近似值在运行时的均值和标准差。
approx = advi.fit(20000, callbacks=[tracker])
Finished [100%]: Average Loss = 2.2862
我们现在可以绘制证据下界和参数迹
fig = plt.figure(figsize=(16, 9))
mu_ax = fig.add_subplot(221)
std_ax = fig.add_subplot(222)
hist_ax = fig.add_subplot(212)
mu_ax.plot(tracker["mean"])
mu_ax.set_title("Mean track")
std_ax.plot(tracker["std"])
std_ax.set_title("Std track")
hist_ax.plot(advi.hist)
hist_ax.set_title("Negative ELBO track");
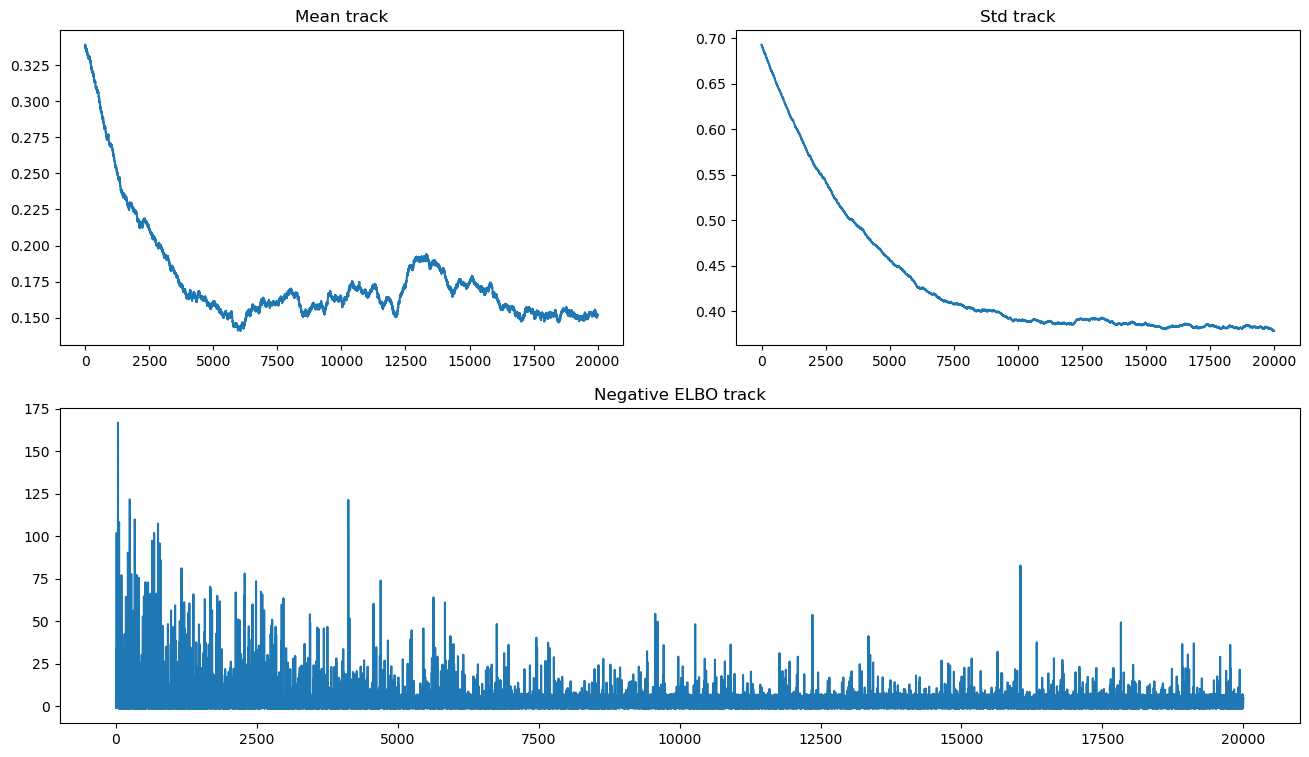
请注意,均值存在收敛问题,并且缺乏收敛似乎不会显着改变 ELBO 轨迹。由于我们正在使用 OO API,我们可以运行更长时间的近似,直到实现收敛。
advi.refine(100_000)
Finished [100%]: Average Loss = 2.1363
让我们看看
fig = plt.figure(figsize=(16, 9))
mu_ax = fig.add_subplot(221)
std_ax = fig.add_subplot(222)
hist_ax = fig.add_subplot(212)
mu_ax.plot(tracker["mean"])
mu_ax.set_title("Mean track")
std_ax.plot(tracker["std"])
std_ax.set_title("Std track")
hist_ax.plot(advi.hist)
hist_ax.set_title("Negative ELBO track");
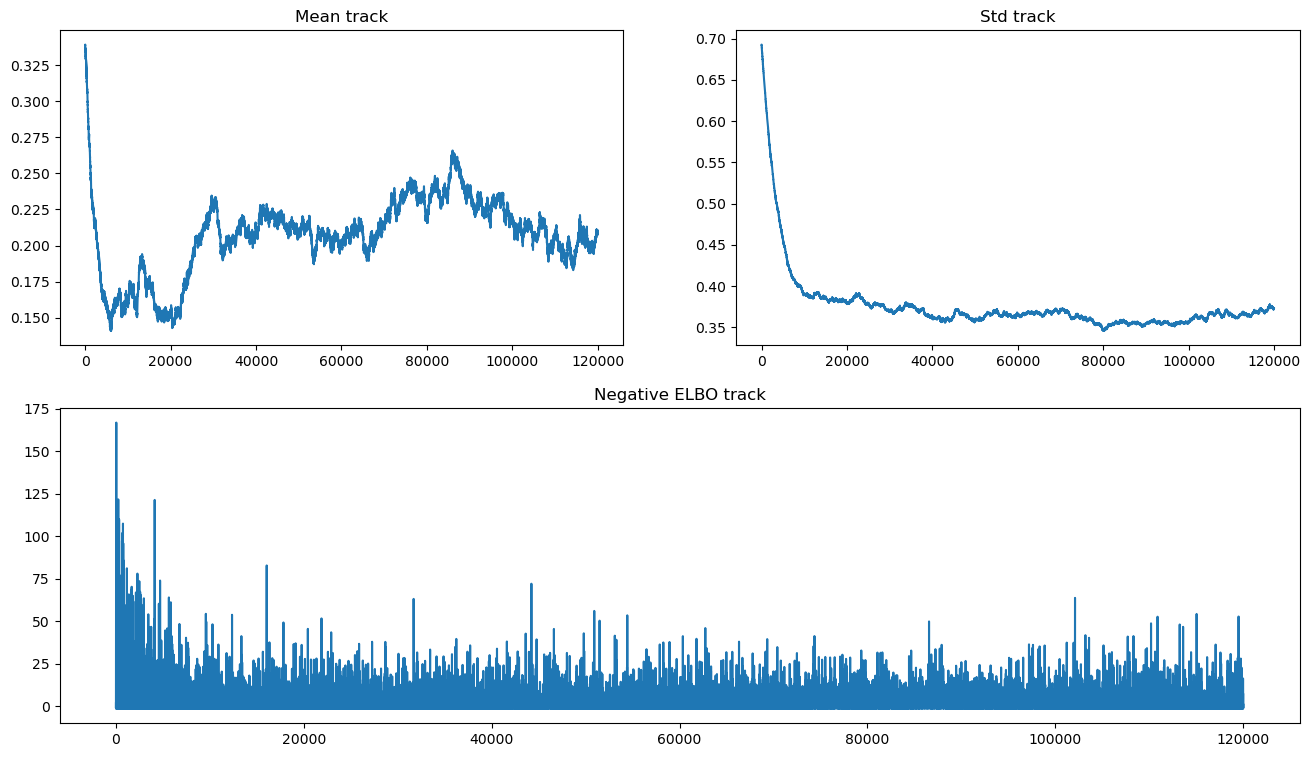
我们仍然看到缺乏收敛的证据,因为均值已退化为随机游走。这可能是选择不良推断算法的结果。无论如何,它是不稳定的,甚至使用不同的随机种子也可能产生非常不同的结果。
让我们将结果与 NUTS 输出进行比较
sns.kdeplot(trace.posterior["x"].values.flatten(), label="NUTS")
sns.kdeplot(approx.sample(20000).posterior["x"].values.flatten(), label="ADVI")
plt.legend();
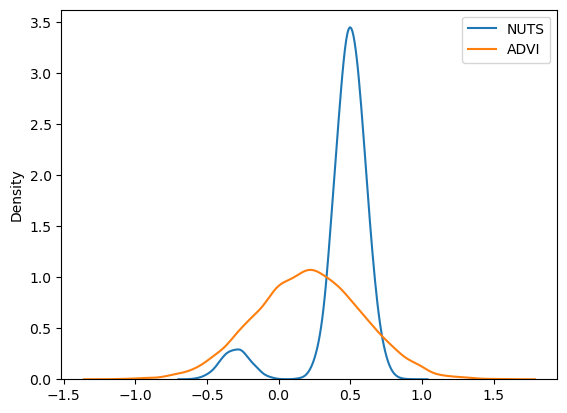
同样,我们看到 ADVI 无法应对多模态;我们可以改用 SVGD,它基于大量粒子生成近似值。
sns.kdeplot(trace.posterior["x"].values.flatten(), label="NUTS")
sns.kdeplot(approx.sample(10000).posterior["x"].values.flatten(), label="ADVI")
sns.kdeplot(svgd_approx.sample(2000).posterior["x"].values.flatten(), label="SVGD")
plt.legend();
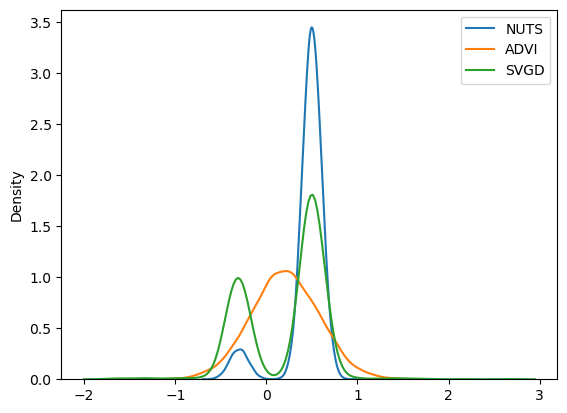
这招奏效了,因为我们现在使用 SVGD 获得了多模态近似。
有了这个,就可以使用这个变分近似计算参数的任意函数。例如,我们可以像使用 NUTS 模型一样计算 \(x^2\) 和 \(sin(x)\)。
# recall x ~ NormalMixture
a = x**2
b = pm.math.sin(x)
为了使用近似值评估这些表达式,我们需要 approx.sample_node。
a_sample = svgd_approx.sample_node(a)
a_sample.eval()
array(0.06251754)
a_sample.eval()
array(0.06251754)
a_sample.eval()
array(0.06251754)
每次调用都会从同一节点产生不同的值。这是因为它具有随机性。
通过应用替换,我们现在摆脱了对 PyMC 模型的依赖;相反,我们现在依赖于近似值。更改它将更改随机节点的分布
有一种更方便的方法可以一次获得大量样本:sample_node
a_samples = svgd_approx.sample_node(a, size=1000)
sns.kdeplot(a_samples.eval())
plt.title("$x^2$ distribution");

sample_node 函数包含一个额外的维度,因此期望或方差的计算由 axis=0 指定。
a_samples.var(0).eval() # variance
array(0.13313996)
a_samples.mean(0).eval() # mean
array(0.24540344)
也可以指定符号样本大小
import pytensor.tensor as pt
i = pt.iscalar("i")
i.tag.test_value = 1
a_samples_i = svgd_approx.sample_node(a, size=i)
a_samples_i.eval({i: 100}).shape
(100,)
a_samples_i.eval({i: 10000}).shape
(10000,)
不幸的是,大小必须是标量值。
多标签逻辑回归#
让我们用著名的 Iris 数据集来说明 Tracker 的用法。我们将尝试多标签分类,并计算预期准确率得分作为诊断。
import pandas as pd
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
X, y = load_iris(return_X_y=True)
X_train, X_test, y_train, y_test = train_test_split(X, y)

一个相对简单的模型在这里就足够了,因为这些类大致是线性可分的;我们将拟合多项逻辑回归。
Xt = pytensor.shared(X_train)
yt = pytensor.shared(y_train)
with pm.Model() as iris_model:
# Coefficients for features
β = pm.Normal("β", 0, sigma=1e2, shape=(4, 3))
# Transoform to unit interval
a = pm.Normal("a", sigma=1e4, shape=(3,))
p = pt.special.softmax(Xt.dot(β) + a, axis=-1)
observed = pm.Categorical("obs", p=p, observed=yt)
在实践中应用替换#
PyMC 模型具有潜在变量的符号输入。为了评估需要了解潜在变量的表达式,需要提供固定值。我们可以为此目的使用 VI 近似的值。sample_node 函数移除符号依赖性。
sample_node 将在每个步骤中使用整个分布,因此我们将在此处使用它。我们可以在单个函数调用中使用 more_replacements 关键字参数在两个替换函数中应用更多替换。
提示: 您也可以在调用
fit时使用more_replacements参数
pm.fit(more_replacements={full_data: minibatch_data})
inference.fit(more_replacements={full_data: minibatch_data})
with iris_model:
# We'll use SVGD
inference = pm.SVGD(n_particles=500, jitter=1)
# Local reference to approximation
approx = inference.approx
# Here we need `more_replacements` to change train_set to test_set
test_probs = approx.sample_node(p, more_replacements={Xt: X_test}, size=100)
# For train set no more replacements needed
train_probs = approx.sample_node(p)
通过应用上面的代码,我们现在为每个观测值获得了 100 个采样的概率(sample_node 的默认数字为 None)。
接下来,我们为采样的准确率得分创建符号表达式
Tracker 期望可调用对象,因此我们可以传递 PyTensor 节点的 .eval 方法,该节点本身就是函数。
对此函数的调用会被缓存,因此可以重复使用。
eval_tracker = pm.callbacks.Tracker(
test_accuracy=test_accuracy.eval, train_accuracy=train_accuracy.eval
)
inference.fit(100, callbacks=[eval_tracker]);
_, ax = plt.subplots(1, 1)
df = pd.DataFrame(eval_tracker["test_accuracy"]).T.melt()
sns.lineplot(x="variable", y="value", data=df, color="red", ax=ax)
ax.plot(eval_tracker["train_accuracy"], color="blue")
ax.set_xlabel("epoch")
plt.legend(["test_accuracy", "train_accuracy"])
plt.title("Training Progress");
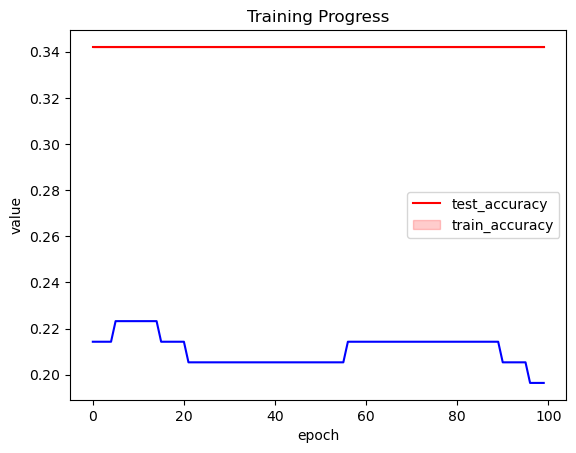
训练似乎在这里不起作用。让我们使用不同的优化器并提高学习率。
inference.fit(400, obj_optimizer=pm.adamax(learning_rate=0.1), callbacks=[eval_tracker]);
_, ax = plt.subplots(1, 1)
df = pd.DataFrame(np.asarray(eval_tracker["test_accuracy"])).T.melt()
sns.lineplot(x="variable", y="value", data=df, color="red", ax=ax)
ax.plot(eval_tracker["train_accuracy"], color="blue")
ax.set_xlabel("epoch")
plt.legend(["test_accuracy", "train_accuracy"])
plt.title("Training Progress");
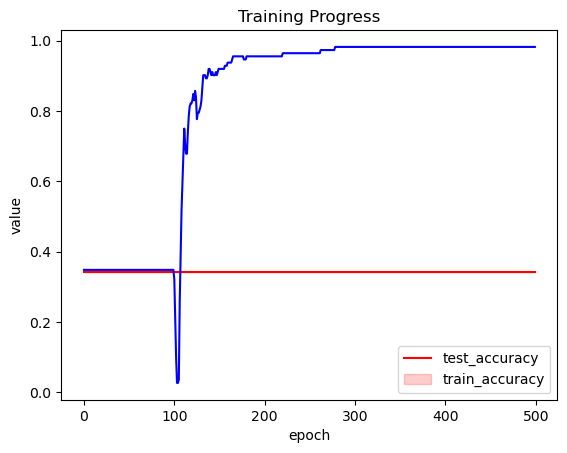
这样好多了!
因此,Tracker 允许我们监视我们的近似值并选择良好的训练计划。
小批量#
当处理大型数据集时,使用小批量训练可以显着加快速度并提高近似性能。大型数据集对梯度的计算施加了沉重的成本。
PyMC 中有一个很好的 API 来处理这些情况,可以通过 pm.Minibatch 类获得。小批量只是一个高度专业化的 PyTensor 张量。
为了演示,让我们模拟大量数据
# Raw values
data = np.random.rand(40000, 100)
# Scaled values
data *= np.random.randint(1, 10, size=(100,))
# Shifted values
data += np.random.rand(100) * 10
为了比较,让我们拟合一个没有小批量处理的模型
with pm.Model() as model:
mu = pm.Flat("mu", shape=(100,))
sd = pm.HalfNormal("sd", shape=(100,))
lik = pm.Normal("lik", mu, sigma=sd, observed=data)
为了好玩,让我们创建一个自定义的特殊用途回调来停止缓慢的优化。这里我们定义一个回调,当近似运行太慢时,它会导致硬停止
def stop_after_10(approx, loss_history, i):
if (i > 0) and (i % 10) == 0:
raise StopIteration("I was slow, sorry")
with model:
advifit = pm.fit(callbacks=[stop_after_10])
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
Cell In[66], line 2
1 with model:
----> 2 advifit = pm.fit(callbacks=[stop_after_10])
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pymc/variational/inference.py:747, in fit(n, method, model, random_seed, start, start_sigma, inf_kwargs, **kwargs)
745 else:
746 raise TypeError(f"method should be one of {set(_select.keys())} or Inference instance")
--> 747 return inference.fit(n, **kwargs)
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pymc/variational/inference.py:138, in Inference.fit(self, n, score, callbacks, progressbar, **kwargs)
136 callbacks = []
137 score = self._maybe_score(score)
--> 138 step_func = self.objective.step_function(score=score, **kwargs)
139 if progressbar:
140 progress = progress_bar(range(n), display=progressbar)
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/configparser.py:47, in _ChangeFlagsDecorator.__call__.<locals>.res(*args, **kwargs)
44 @wraps(f)
45 def res(*args, **kwargs):
46 with self:
---> 47 return f(*args, **kwargs)
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pymc/variational/opvi.py:387, in ObjectiveFunction.step_function(self, obj_n_mc, tf_n_mc, obj_optimizer, test_optimizer, more_obj_params, more_tf_params, more_updates, more_replacements, total_grad_norm_constraint, score, fn_kwargs)
385 seed = self.approx.rng.randint(2**30, dtype=np.int64)
386 if score:
--> 387 step_fn = compile_pymc([], updates.loss, updates=updates, random_seed=seed, **fn_kwargs)
388 else:
389 step_fn = compile_pymc([], [], updates=updates, random_seed=seed, **fn_kwargs)
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pymc/pytensorf.py:1121, in compile_pymc(inputs, outputs, random_seed, mode, **kwargs)
1119 opt_qry = mode.provided_optimizer.including("random_make_inplace", check_parameter_opt)
1120 mode = Mode(linker=mode.linker, optimizer=opt_qry)
-> 1121 pytensor_function = pytensor.function(
1122 inputs,
1123 outputs,
1124 updates={**rng_updates, **kwargs.pop("updates", {})},
1125 mode=mode,
1126 **kwargs,
1127 )
1128 return pytensor_function
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/compile/function/__init__.py:315, in function(inputs, outputs, mode, updates, givens, no_default_updates, accept_inplace, name, rebuild_strict, allow_input_downcast, profile, on_unused_input)
309 fn = orig_function(
310 inputs, outputs, mode=mode, accept_inplace=accept_inplace, name=name
311 )
312 else:
313 # note: pfunc will also call orig_function -- orig_function is
314 # a choke point that all compilation must pass through
--> 315 fn = pfunc(
316 params=inputs,
317 outputs=outputs,
318 mode=mode,
319 updates=updates,
320 givens=givens,
321 no_default_updates=no_default_updates,
322 accept_inplace=accept_inplace,
323 name=name,
324 rebuild_strict=rebuild_strict,
325 allow_input_downcast=allow_input_downcast,
326 on_unused_input=on_unused_input,
327 profile=profile,
328 output_keys=output_keys,
329 )
330 return fn
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/compile/function/pfunc.py:367, in pfunc(params, outputs, mode, updates, givens, no_default_updates, accept_inplace, name, rebuild_strict, allow_input_downcast, profile, on_unused_input, output_keys, fgraph)
353 profile = ProfileStats(message=profile)
355 inputs, cloned_outputs = construct_pfunc_ins_and_outs(
356 params,
357 outputs,
(...)
364 fgraph=fgraph,
365 )
--> 367 return orig_function(
368 inputs,
369 cloned_outputs,
370 mode,
371 accept_inplace=accept_inplace,
372 name=name,
373 profile=profile,
374 on_unused_input=on_unused_input,
375 output_keys=output_keys,
376 fgraph=fgraph,
377 )
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/compile/function/types.py:1766, in orig_function(inputs, outputs, mode, accept_inplace, name, profile, on_unused_input, output_keys, fgraph)
1754 m = Maker(
1755 inputs,
1756 outputs,
(...)
1763 fgraph=fgraph,
1764 )
1765 with config.change_flags(compute_test_value="off"):
-> 1766 fn = m.create(defaults)
1767 finally:
1768 t2 = time.perf_counter()
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/compile/function/types.py:1659, in FunctionMaker.create(self, input_storage, trustme, storage_map)
1656 start_import_time = pytensor.link.c.cmodule.import_time
1658 with config.change_flags(traceback__limit=config.traceback__compile_limit):
-> 1659 _fn, _i, _o = self.linker.make_thunk(
1660 input_storage=input_storage_lists, storage_map=storage_map
1661 )
1663 end_linker = time.perf_counter()
1665 linker_time = end_linker - start_linker
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/link/basic.py:254, in LocalLinker.make_thunk(self, input_storage, output_storage, storage_map, **kwargs)
247 def make_thunk(
248 self,
249 input_storage: Optional["InputStorageType"] = None,
(...)
252 **kwargs,
253 ) -> Tuple["BasicThunkType", "InputStorageType", "OutputStorageType"]:
--> 254 return self.make_all(
255 input_storage=input_storage,
256 output_storage=output_storage,
257 storage_map=storage_map,
258 )[:3]
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/link/vm.py:1246, in VMLinker.make_all(self, profiler, input_storage, output_storage, storage_map)
1241 thunk_start = time.perf_counter()
1242 # no-recycling is done at each VM.__call__ So there is
1243 # no need to cause duplicate c code by passing
1244 # no_recycling here.
1245 thunks.append(
-> 1246 node.op.make_thunk(node, storage_map, compute_map, [], impl=impl)
1247 )
1248 linker_make_thunk_time[node] = time.perf_counter() - thunk_start
1249 if not hasattr(thunks[-1], "lazy"):
1250 # We don't want all ops maker to think about lazy Ops.
1251 # So if they didn't specify that its lazy or not, it isn't.
1252 # If this member isn't present, it will crash later.
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/link/c/op.py:131, in COp.make_thunk(self, node, storage_map, compute_map, no_recycling, impl)
127 self.prepare_node(
128 node, storage_map=storage_map, compute_map=compute_map, impl="c"
129 )
130 try:
--> 131 return self.make_c_thunk(node, storage_map, compute_map, no_recycling)
132 except (NotImplementedError, MethodNotDefined):
133 # We requested the c code, so don't catch the error.
134 if impl == "c":
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/link/c/op.py:96, in COp.make_c_thunk(self, node, storage_map, compute_map, no_recycling)
94 print(f"Disabling C code for {self} due to unsupported float16")
95 raise NotImplementedError("float16")
---> 96 outputs = cl.make_thunk(
97 input_storage=node_input_storage, output_storage=node_output_storage
98 )
99 thunk, node_input_filters, node_output_filters = outputs
101 @is_cthunk_wrapper_type
102 def rval():
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/link/c/basic.py:1202, in CLinker.make_thunk(self, input_storage, output_storage, storage_map, cache, **kwargs)
1167 """Compile this linker's `self.fgraph` and return a function that performs the computations.
1168
1169 The return values can be used as follows:
(...)
1199
1200 """
1201 init_tasks, tasks = self.get_init_tasks()
-> 1202 cthunk, module, in_storage, out_storage, error_storage = self.__compile__(
1203 input_storage, output_storage, storage_map, cache
1204 )
1206 res = _CThunk(cthunk, init_tasks, tasks, error_storage, module)
1207 res.nodes = self.node_order
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/link/c/basic.py:1122, in CLinker.__compile__(self, input_storage, output_storage, storage_map, cache)
1120 input_storage = tuple(input_storage)
1121 output_storage = tuple(output_storage)
-> 1122 thunk, module = self.cthunk_factory(
1123 error_storage,
1124 input_storage,
1125 output_storage,
1126 storage_map,
1127 cache,
1128 )
1129 return (
1130 thunk,
1131 module,
(...)
1140 error_storage,
1141 )
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/link/c/basic.py:1647, in CLinker.cthunk_factory(self, error_storage, in_storage, out_storage, storage_map, cache)
1645 if cache is None:
1646 cache = get_module_cache()
-> 1647 module = cache.module_from_key(key=key, lnk=self)
1649 vars = self.inputs + self.outputs + self.orphans
1650 # List of indices that should be ignored when passing the arguments
1651 # (basically, everything that the previous call to uniq eliminated)
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/link/c/cmodule.py:1231, in ModuleCache.module_from_key(self, key, lnk)
1229 try:
1230 location = dlimport_workdir(self.dirname)
-> 1231 module = lnk.compile_cmodule(location)
1232 name = module.__file__
1233 assert name.startswith(location)
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/link/c/basic.py:1546, in CLinker.compile_cmodule(self, location)
1544 try:
1545 _logger.debug(f"LOCATION {location}")
-> 1546 module = c_compiler.compile_str(
1547 module_name=mod.code_hash,
1548 src_code=src_code,
1549 location=location,
1550 include_dirs=self.header_dirs(),
1551 lib_dirs=self.lib_dirs(),
1552 libs=libs,
1553 preargs=preargs,
1554 )
1555 except Exception as e:
1556 e.args += (str(self.fgraph),)
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/link/c/cmodule.py:2591, in GCC_compiler.compile_str(module_name, src_code, location, include_dirs, lib_dirs, libs, preargs, py_module, hide_symbols)
2588 print(" ".join(cmd), file=sys.stderr)
2590 try:
-> 2591 p_out = output_subprocess_Popen(cmd)
2592 compile_stderr = p_out[1].decode()
2593 except Exception:
2594 # An exception can occur e.g. if `g++` is not found.
File ~/mambaforge/envs/pie/lib/python3.9/site-packages/pytensor/utils.py:261, in output_subprocess_Popen(command, **params)
258 p = subprocess_Popen(command, **params)
259 # we need to use communicate to make sure we don't deadlock around
260 # the stdout/stderr pipe.
--> 261 out = p.communicate()
262 return out + (p.returncode,)
File ~/mambaforge/envs/pie/lib/python3.9/subprocess.py:1130, in Popen.communicate(self, input, timeout)
1127 endtime = None
1129 try:
-> 1130 stdout, stderr = self._communicate(input, endtime, timeout)
1131 except KeyboardInterrupt:
1132 # https://bugs.python.org/issue25942
1133 # See the detailed comment in .wait().
1134 if timeout is not None:
File ~/mambaforge/envs/pie/lib/python3.9/subprocess.py:1977, in Popen._communicate(self, input, endtime, orig_timeout)
1970 self._check_timeout(endtime, orig_timeout,
1971 stdout, stderr,
1972 skip_check_and_raise=True)
1973 raise RuntimeError( # Impossible :)
1974 '_check_timeout(..., skip_check_and_raise=True) '
1975 'failed to raise TimeoutExpired.')
-> 1977 ready = selector.select(timeout)
1978 self._check_timeout(endtime, orig_timeout, stdout, stderr)
1980 # XXX Rewrite these to use non-blocking I/O on the file
1981 # objects; they are no longer using C stdio!
File ~/mambaforge/envs/pie/lib/python3.9/selectors.py:416, in _PollLikeSelector.select(self, timeout)
414 ready = []
415 try:
--> 416 fd_event_list = self._selector.poll(timeout)
417 except InterruptedError:
418 return ready
KeyboardInterrupt:
推断太慢了,每次迭代需要几秒钟;拟合近似值将花费数小时!
现在让我们使用小批量。在每次迭代中,我们将抽取 500 个随机值
记住在 observed 中设置
total_size
total_size 是一个重要的参数,它允许 PyMC 推断正确的密度重缩放方式。如果未设置,您可能会得到完全错误的结果。有关更多信息,请参阅 pm.Minibatch 的完整文档。
X = pm.Minibatch(data, batch_size=500)
with pm.Model() as model:
mu = pm.Normal("mu", 0, sigma=1e5, shape=(100,))
sd = pm.HalfNormal("sd", shape=(100,))
likelihood = pm.Normal("likelihood", mu, sigma=sd, observed=X, total_size=data.shape)
with model:
advifit = pm.fit()
Finished [100%]: Average Loss = 1.5101e+05
plt.plot(advifit.hist);
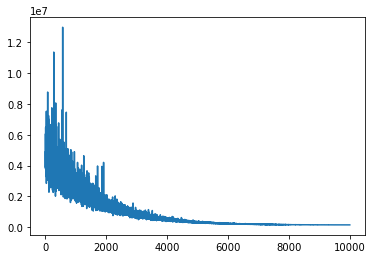
小批量推断速度显着加快。在某些需要进行矩阵分解或模型非常宽的情况下,可能需要多维小批量。
这是 Minibatch 的文档字符串,用于说明如何自定义它。
print(pm.Minibatch.__doc__)
Multidimensional minibatch that is pure TensorVariable
Parameters
----------
data: np.ndarray
initial data
batch_size: ``int`` or ``List[int|tuple(size, random_seed)]``
batch size for inference, random seed is needed
for child random generators
dtype: ``str``
cast data to specific type
broadcastable: tuple[bool]
change broadcastable pattern that defaults to ``(False, ) * ndim``
name: ``str``
name for tensor, defaults to "Minibatch"
random_seed: ``int``
random seed that is used by default
update_shared_f: ``callable``
returns :class:`ndarray` that will be carefully
stored to underlying shared variable
you can use it to change source of
minibatches programmatically
in_memory_size: ``int`` or ``List[int|slice|Ellipsis]``
data size for storing in ``aesara.shared``
Attributes
----------
shared: shared tensor
Used for storing data
minibatch: minibatch tensor
Used for training
Notes
-----
Below is a common use case of Minibatch with variational inference.
Importantly, we need to make PyMC "aware" that a minibatch is being used in inference.
Otherwise, we will get the wrong :math:`logp` for the model.
the density of the model ``logp`` that is affected by Minibatch. See more in the examples below.
To do so, we need to pass the ``total_size`` parameter to the observed node, which correctly scales
the density of the model ``logp`` that is affected by Minibatch. See more in the examples below.
Examples
--------
Consider we have `data` as follows:
>>> data = np.random.rand(100, 100)
if we want a 1d slice of size 10 we do
>>> x = Minibatch(data, batch_size=10)
Note that your data is cast to ``floatX`` if it is not integer type
But you still can add the ``dtype`` kwarg for :class:`Minibatch`
if you need more control.
If we want 10 sampled rows and columns
``[(size, seed), (size, seed)]`` we can use
>>> x = Minibatch(data, batch_size=[(10, 42), (10, 42)], dtype='int32')
>>> assert str(x.dtype) == 'int32'
Or, more simply, we can use the default random seed = 42
``[size, size]``
>>> x = Minibatch(data, batch_size=[10, 10])
In the above, `x` is a regular :class:`TensorVariable` that supports any math operations:
>>> assert x.eval().shape == (10, 10)
You can pass the Minibatch `x` to your desired model:
>>> with pm.Model() as model:
... mu = pm.Flat('mu')
... sigma = pm.HalfNormal('sigma')
... lik = pm.Normal('lik', mu, sigma, observed=x, total_size=(100, 100))
Then you can perform regular Variational Inference out of the box
>>> with model:
... approx = pm.fit()
Important note: :class:``Minibatch`` has ``shared``, and ``minibatch`` attributes
you can call later:
>>> x.set_value(np.random.laplace(size=(100, 100)))
and minibatches will be then from new storage
it directly affects ``x.shared``.
A less convenient convenient, but more explicit, way to achieve the same
thing:
>>> x.shared.set_value(pm.floatX(np.random.laplace(size=(100, 100))))
The programmatic way to change storage is as follows
I import ``partial`` for simplicity
>>> from functools import partial
>>> datagen = partial(np.random.laplace, size=(100, 100))
>>> x = Minibatch(datagen(), batch_size=10, update_shared_f=datagen)
>>> x.update_shared()
To be more concrete about how we create a minibatch, here is a demo:
1. create a shared variable
>>> shared = aesara.shared(data)
2. take a random slice of size 10:
>>> ridx = pm.at_rng().uniform(size=(10,), low=0, high=data.shape[0]-1e-10).astype('int64')
3) take the resulting slice:
>>> minibatch = shared[ridx]
That's done. Now you can use this minibatch somewhere else.
You can see that the implementation does not require a fixed shape
for the shared variable. Feel free to use that if needed.
*FIXME: What is "that" which we can use here? A fixed shape? Should this say
"but feel free to put a fixed shape on the shared variable, if appropriate?"*
Suppose you need to make some replacements in the graph, e.g. change the minibatch to testdata
>>> node = x ** 2 # arbitrary expressions on minibatch `x`
>>> testdata = pm.floatX(np.random.laplace(size=(1000, 10)))
Then you should create a `dict` with replacements:
>>> replacements = {x: testdata}
>>> rnode = aesara.clone_replace(node, replacements)
>>> assert (testdata ** 2 == rnode.eval()).all()
*FIXME: In the following, what is the **reason** to replace the Minibatch variable with
its shared variable? And in the following, the `rnode` is a **new** node, not a modification
of a previously existing node, correct?*
To replace a minibatch with its shared variable you should do
the same things. The Minibatch variable is accessible through the `minibatch` attribute.
For example
>>> replacements = {x.minibatch: x.shared}
>>> rnode = aesara.clone_replace(node, replacements)
For more complex slices some more code is needed that can seem not so clear
>>> moredata = np.random.rand(10, 20, 30, 40, 50)
The default ``total_size`` that can be passed to PyMC random node
is then ``(10, 20, 30, 40, 50)`` but can be less verbose in some cases
1. Advanced indexing, ``total_size = (10, Ellipsis, 50)``
>>> x = Minibatch(moredata, [2, Ellipsis, 10])
We take the slice only for the first and last dimension
>>> assert x.eval().shape == (2, 20, 30, 40, 10)
2. Skipping a particular dimension, ``total_size = (10, None, 30)``:
>>> x = Minibatch(moredata, [2, None, 20])
>>> assert x.eval().shape == (2, 20, 20, 40, 50)
3. Mixing both of these together, ``total_size = (10, None, 30, Ellipsis, 50)``:
>>> x = Minibatch(moredata, [2, None, 20, Ellipsis, 10])
>>> assert x.eval().shape == (2, 20, 20, 40, 10)
水印#
%load_ext watermark
%watermark -n -u -v -iv -w
Last updated: Sun Nov 20 2022
Python implementation: CPython
Python version : 3.10.4
IPython version : 8.4.0
sys : 3.10.4 | packaged by conda-forge | (main, Mar 24 2022, 17:39:37) [Clang 12.0.1 ]
pymc : 4.3.0
seaborn : 0.11.2
arviz : 0.13.0
numpy : 1.22.4
matplotlib: 3.5.2
aesara : 2.8.9+11.ge8eed6c18
pandas : 1.4.2
Watermark: 2.3.1
许可声明#
此示例库中的所有 Notebook 均根据 MIT 许可证提供,该许可证允许修改和再分发以用于任何用途,前提是保留版权和许可声明。
引用 PyMC 示例#
要引用此 Notebook,请使用 Zenodo 为 pymc-examples 存储库提供的 DOI。
重要提示
许多 Notebook 改编自其他来源:博客、书籍…… 在这种情况下,您也应该引用原始来源。
另请记住引用您的代码使用的相关库。
这是一个 bibtex 格式的引用模板
@incollection{citekey,
author = "<notebook authors, see above>",
title = "<notebook title>",
editor = "PyMC Team",
booktitle = "PyMC examples",
doi = "10.5281/zenodo.5654871"
}
渲染后可能看起来像这样
