ODE Lotka-Volterra 模型贝叶斯推断的多种方法#
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pymc as pm
import pytensor
import pytensor.tensor as pt
from numba import njit
from pymc.ode import DifferentialEquation
from pytensor.compile.ops import as_op
from scipy.integrate import odeint
from scipy.optimize import least_squares
print(f"Running on PyMC v{pm.__version__}")
Running on PyMC v5.1.2+24.gf3ce16f26
%load_ext watermark
az.style.use("arviz-darkgrid")
rng = np.random.default_rng(1234)
目的#
本笔记本的目的是演示如何在常微分方程 (ODE) 系统上执行贝叶斯推断,包括有梯度和无梯度两种情况。 并比较不同采样器的准确性和效率。
我们将首先介绍 Lotka-Volterra 捕食者-猎物 ODE 模型和示例数据。 接下来,我们将使用 scipy.odeint 和(非贝叶斯)最小二乘优化来求解 ODE。 接下来,我们在 PyMC 中使用无梯度采样器执行贝叶斯推断。 最后,我们使用基于梯度的采样器并比较结果。
主要结论#
根据本笔记本中的实验,对 Lotka-Volterra 方程执行贝叶斯推断最简单有效的方法是在 Scipy 中指定 ODE 系统,将该函数包装为 Pytensor op,并在 PyMC 中使用微分进化 Metropolis (DEMetropolis) 采样器。
背景#
动机#
常微分方程模型 (ODE) 广泛应用于科学和工程领域,用于模拟物理变量的时间演化。 给定实验数据,估计模型参数的值和不确定性的自然选择是贝叶斯推断。 然而,ODE 在贝叶斯设置中可能难以指定和求解,因此,本笔记本逐步介绍了使用 PyMC 解决 ODE 推断问题的多种方法。 本例中使用的 Lotka-Volterra 模型经常被用于贝叶斯推断方法的基准测试(例如,在 Stan 案例研究中,以及 *Statistical Rethinking* 的第 16 章 [McElreath, 2018]中)。
Lotka-Volterra 捕食者-猎物模型#
Lotka-Volterra 模型描述了捕食者和猎物物种之间的相互作用。 此 ODE 由下式给出
状态向量 \(X(t)=[x(t),y(t)]\) 分别包含猎物和捕食者物种的密度。 参数 \(\boldsymbol{\theta}=[\alpha,\beta,\gamma,\delta, x(0),y(0)]\) 是我们希望从实验观察中推断出的未知数。 \(x(0), y(0)\) 是求解 ODE 所需的状态初始值,而 \(\alpha,\beta,\gamma\) 和 \(\delta\) 是未知模型参数,它们代表以下含义
\(\alpha\) 是没有捕食者时猎物的增长率。
\(\beta\) 是猎物因捕食而死亡的死亡率。
\(\gamma\) 是没有猎物时捕食者的死亡率。
\(\delta\) 是在猎物存在的情况下捕食者的增长率。
哈德逊湾公司数据#
Lotka-Volterra 捕食者猎物模型已成功用于解释自然界中捕食者和猎物种群的动态,例如哈德逊湾公司的猞猁和雪兔数据。 由于数据集很小,我们将手动输入这些值。
# fmt: off
data = pd.DataFrame(dict(
year = np.arange(1900., 1921., 1),
lynx = np.array([4.0, 6.1, 9.8, 35.2, 59.4, 41.7, 19.0, 13.0, 8.3, 9.1, 7.4,
8.0, 12.3, 19.5, 45.7, 51.1, 29.7, 15.8, 9.7, 10.1, 8.6]),
hare = np.array([30.0, 47.2, 70.2, 77.4, 36.3, 20.6, 18.1, 21.4, 22.0, 25.4,
27.1, 40.3, 57.0, 76.6, 52.3, 19.5, 11.2, 7.6, 14.6, 16.2, 24.7])))
data.head()
# fmt: on
| 年份 | 猞猁 | 兔 | |
|---|---|---|---|
| 0 | 1900.0 | 4.0 | 30.0 |
| 1 | 1901.0 | 6.1 | 47.2 |
| 2 | 1902.0 | 9.8 | 70.2 |
| 3 | 1903.0 | 35.2 | 77.4 |
| 4 | 1904.0 | 59.4 | 36.3 |
# plot data function for reuse later
def plot_data(ax, lw=2, title="Hudson's Bay Company Data"):
ax.plot(data.year, data.lynx, color="b", lw=lw, marker="o", markersize=12, label="Lynx (Data)")
ax.plot(data.year, data.hare, color="g", lw=lw, marker="+", markersize=14, label="Hare (Data)")
ax.legend(fontsize=14, loc="center left", bbox_to_anchor=(1, 0.5))
ax.set_xlim([1900, 1920])
ax.set_ylim(0)
ax.set_xlabel("Year", fontsize=14)
ax.set_ylabel("Pelts (Thousands)", fontsize=14)
ax.set_xticks(data.year.astype(int))
ax.set_xticklabels(ax.get_xticks(), rotation=45)
ax.set_title(title, fontsize=16)
return ax
_, ax = plt.subplots(figsize=(12, 4))
plot_data(ax);

问题陈述#
本分析的目的是有不确定性地估计 1900 年至 1920 年哈德逊湾公司数据的 Lotka-Volterra 模型的参数。
Scipy odeint#
在这里,我们制作一个 Python 函数来表示 ODE 方程的右侧,其调用签名是 odeint 函数所需的。 请注意,Scipy 的 solve_ivp 也可以使用,但在速度测试中,较旧的 odeint 函数速度更快,因此在本笔记本中使用。
# define the right hand side of the ODE equations in the Scipy odeint signature
from numba import njit
@njit
def rhs(X, t, theta):
# unpack parameters
x, y = X
alpha, beta, gamma, delta, xt0, yt0 = theta
# equations
dx_dt = alpha * x - beta * x * y
dy_dt = -gamma * y + delta * x * y
return [dx_dt, dy_dt]
为了了解模型并确保方程正常工作,让我们使用 \(\theta\) 的合理值运行模型一次,并绘制结果。
# plot model function
def plot_model(
ax,
x_y,
time=np.arange(1900, 1921, 0.01),
alpha=1,
lw=3,
title="Hudson's Bay Company Data and\nExample Model Run",
):
ax.plot(time, x_y[:, 1], color="b", alpha=alpha, lw=lw, label="Lynx (Model)")
ax.plot(time, x_y[:, 0], color="g", alpha=alpha, lw=lw, label="Hare (Model)")
ax.legend(fontsize=14, loc="center left", bbox_to_anchor=(1, 0.5))
ax.set_title(title, fontsize=16)
return ax
# note theta = alpha, beta, gamma, delta, xt0, yt0
theta = np.array([0.52, 0.026, 0.84, 0.026, 34.0, 5.9])
time = np.arange(1900, 1921, 0.01)
# call Scipy's odeint function
x_y = odeint(func=rhs, y0=theta[-2:], t=time, args=(theta,))
# plot
_, ax = plt.subplots(figsize=(12, 4))
plot_data(ax, lw=0)
plot_model(ax, x_y);
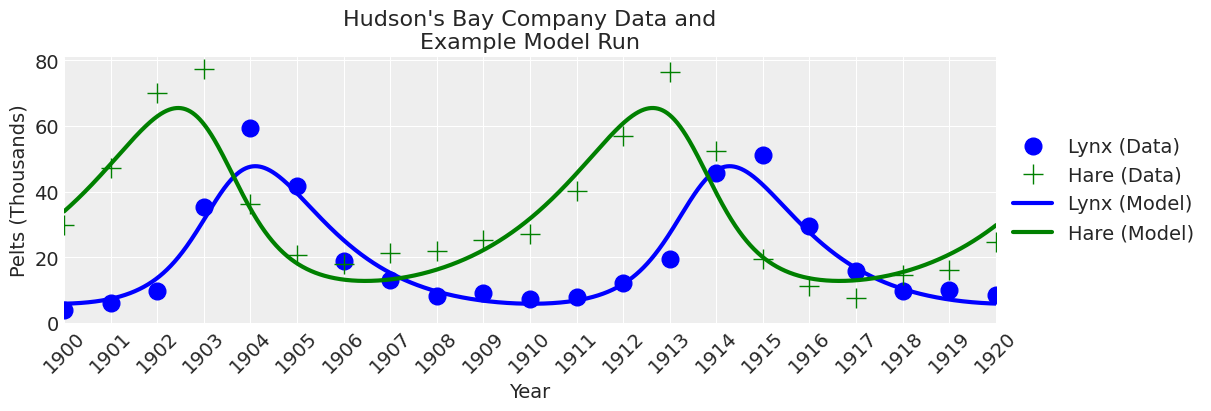
看起来 odeint 函数运行正常。
最小二乘解#
现在,我们可以使用最小二乘法求解 ODE。 制作一个计算残差的函数。
# function that calculates residuals based on a given theta
def ode_model_resid(theta):
return (
data[["hare", "lynx"]] - odeint(func=rhs, y0=theta[-2:], t=data.year, args=(theta,))
).values.flatten()
将残差函数馈送到 Scipy least_squares 求解器。
# calculate least squares using the Scipy solver
results = least_squares(ode_model_resid, x0=theta)
# put the results in a dataframe for presentation and convenience
df = pd.DataFrame()
parameter_names = ["alpha", "beta", "gamma", "delta", "h0", "l0"]
df["Parameter"] = parameter_names
df["Least Squares Solution"] = results.x
df.round(2)
| 参数 | 最小二乘解 | |
|---|---|---|
| 0 | alpha | 0.48 |
| 1 | beta | 0.02 |
| 2 | gamma | 0.93 |
| 3 | delta | 0.03 |
| 4 | h0 | 34.91 |
| 5 | l0 | 3.86 |
绘图
time = np.arange(1900, 1921, 0.01)
theta = results.x
x_y = odeint(func=rhs, y0=theta[-2:], t=time, args=(theta,))
fig, ax = plt.subplots(figsize=(12, 4))
plot_data(ax, lw=0)
plot_model(ax, x_y, title="Least Squares Solution");
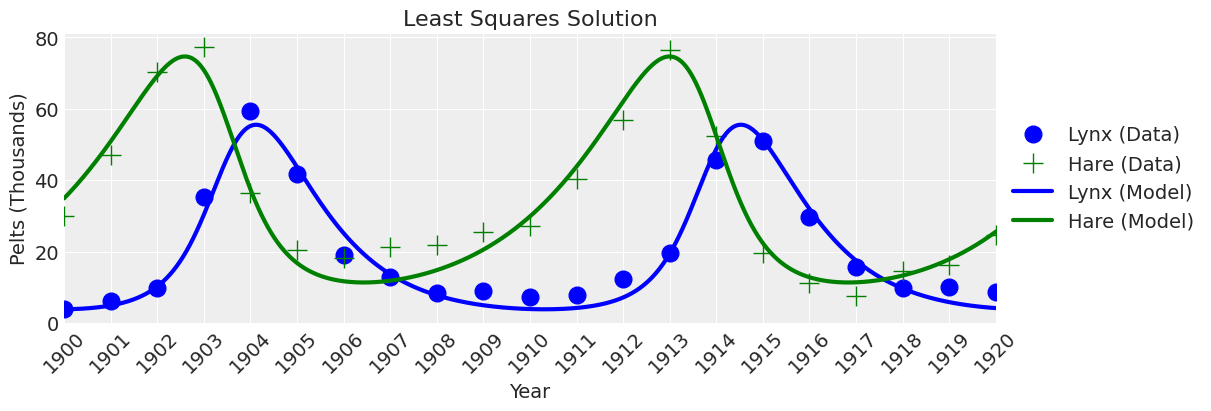
看起来不错。 如果我们不关心不确定性,那么我们就完成了。 但我们确实关心不确定性,所以让我们继续进行贝叶斯推断。
用于无梯度贝叶斯推断的 PyMC 模型规范#
与其他基于 Numpy 或 Scipy 的函数一样,scipy.integrate.odeint 函数不能直接在 PyMC 模型中使用,因为 PyMC 需要知道变量的输入和输出类型才能编译。 因此,我们使用 Pytensor 包装器为 PyMC 提供变量类型。 然后,该函数可以与无梯度采样器结合在 PyMC 中使用。
使用 @as_op 装饰器将 Python 函数转换为 Pytensor 运算符#
我们使用 @as_op 装饰器告诉 PyMC 输入变量类型和输出变量类型。 odeint 返回 Numpy 数组,但为此目的,我们告诉 PyMC 它们是 Pytensor 双精度浮点张量。
PyMC 模型#
现在,我们可以使用 ODE 求解器指定 PyMC 模型! 对于先验,我们将使用最小二乘计算的结果 (results.x) 来分配从正确范围开始的先验。 这些是经验导出的弱信息先验。 我们还使它们仅对这个问题为正。
我们将对未转换的数据(即,未对数转换)使用正态似然,以最好地拟合数据的峰值。
theta = results.x # least squares solution used to inform the priors
with pm.Model() as model:
# Priors
alpha = pm.TruncatedNormal("alpha", mu=theta[0], sigma=0.1, lower=0, initval=theta[0])
beta = pm.TruncatedNormal("beta", mu=theta[1], sigma=0.01, lower=0, initval=theta[1])
gamma = pm.TruncatedNormal("gamma", mu=theta[2], sigma=0.1, lower=0, initval=theta[2])
delta = pm.TruncatedNormal("delta", mu=theta[3], sigma=0.01, lower=0, initval=theta[3])
xt0 = pm.TruncatedNormal("xto", mu=theta[4], sigma=1, lower=0, initval=theta[4])
yt0 = pm.TruncatedNormal("yto", mu=theta[5], sigma=1, lower=0, initval=theta[5])
sigma = pm.HalfNormal("sigma", 10)
# Ode solution function
ode_solution = pytensor_forward_model_matrix(
pm.math.stack([alpha, beta, gamma, delta, xt0, yt0])
)
# Likelihood
pm.Normal("Y_obs", mu=ode_solution, sigma=sigma, observed=data[["hare", "lynx"]].values)
pm.model_to_graphviz(model=model)
绘图函数#
一些我们将在下面重用的绘图函数。
def plot_model_trace(ax, trace_df, row_idx, lw=1, alpha=0.2):
cols = ["alpha", "beta", "gamma", "delta", "xto", "yto"]
row = trace_df.iloc[row_idx, :][cols].values
# alpha, beta, gamma, delta, Xt0, Yt0
time = np.arange(1900, 1921, 0.01)
theta = row
x_y = odeint(func=rhs, y0=theta[-2:], t=time, args=(theta,))
plot_model(ax, x_y, time=time, lw=lw, alpha=alpha);
def plot_inference(
ax,
trace,
num_samples=25,
title="Hudson's Bay Company Data and\nInference Model Runs",
plot_model_kwargs=dict(lw=1, alpha=0.2),
):
trace_df = az.extract(trace, num_samples=num_samples).to_dataframe()
plot_data(ax, lw=0)
for row_idx in range(num_samples):
plot_model_trace(ax, trace_df, row_idx, **plot_model_kwargs)
handles, labels = ax.get_legend_handles_labels()
ax.legend(handles[:2], labels[:2], loc="center left", bbox_to_anchor=(1, 0.5))
ax.set_title(title, fontsize=16)
无梯度采样器选项#
拥有良好的无梯度采样器可以打开可以在 PyMC 中拟合的模型。 PyMC 中有五个适用于此问题的无梯度采样器选项
Slice- 默认的无梯度采样器DEMetropolisZ- 一种微分进化 Metropolis 采样器,它使用过去的信息来通知采样跳跃DEMetropolis- 一种微分进化 Metropolis 采样器Metropolis- vanilla Metropolis 采样器SMC- 顺序蒙特卡洛
让我们试一试。
关于运行这些推断的一些注意事项。 对于每个采样器,调整步骤和抽取数量都已减少,以便在合理的时间内(分钟级)运行推断。 这不足以获得良好的推断,在某些情况下,但这对于演示目的来说是可行的。 此外,多核处理不适用于所有机器上的 Pytensor op 函数,因此推断是在一个核心上执行的。
Slice 采样器#
# Variable list to give to the sample step parameter
vars_list = list(model.values_to_rvs.keys())[:-1]
# Specify the sampler
sampler = "Slice Sampler"
tune = draws = 2000
# Inference!
with model:
trace_slice = pm.sample(step=[pm.Slice(vars_list)], tune=tune, draws=draws)
trace = trace_slice
az.summary(trace)
Multiprocess sampling (4 chains in 4 jobs)
CompoundStep
>Slice: [alpha]
>Slice: [beta]
>Slice: [gamma]
>Slice: [delta]
>Slice: [xto]
>Slice: [yto]
>Slice: [sigma]
Sampling 4 chains for 2_000 tune and 2_000 draw iterations (8_000 + 8_000 draws total) took 120 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for details
| 均值 | 标准差 | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 0.478 | 0.025 | 0.433 | 0.526 | 0.002 | 0.002 | 115.0 | 254.0 | 1.04 |
| beta | 0.025 | 0.001 | 0.022 | 0.027 | 0.000 | 0.000 | 253.0 | 497.0 | 1.01 |
| gamma | 0.937 | 0.054 | 0.835 | 1.039 | 0.005 | 0.004 | 109.0 | 241.0 | 1.04 |
| delta | 0.028 | 0.002 | 0.025 | 0.031 | 0.000 | 0.000 | 109.0 | 242.0 | 1.05 |
| xto | 34.945 | 0.823 | 33.386 | 36.472 | 0.023 | 0.016 | 1269.0 | 2646.0 | 1.00 |
| yto | 3.837 | 0.476 | 2.958 | 4.730 | 0.036 | 0.026 | 169.0 | 491.0 | 1.03 |
| sigma | 4.111 | 0.487 | 3.263 | 5.038 | 0.007 | 0.005 | 5141.0 | 5579.0 | 1.00 |
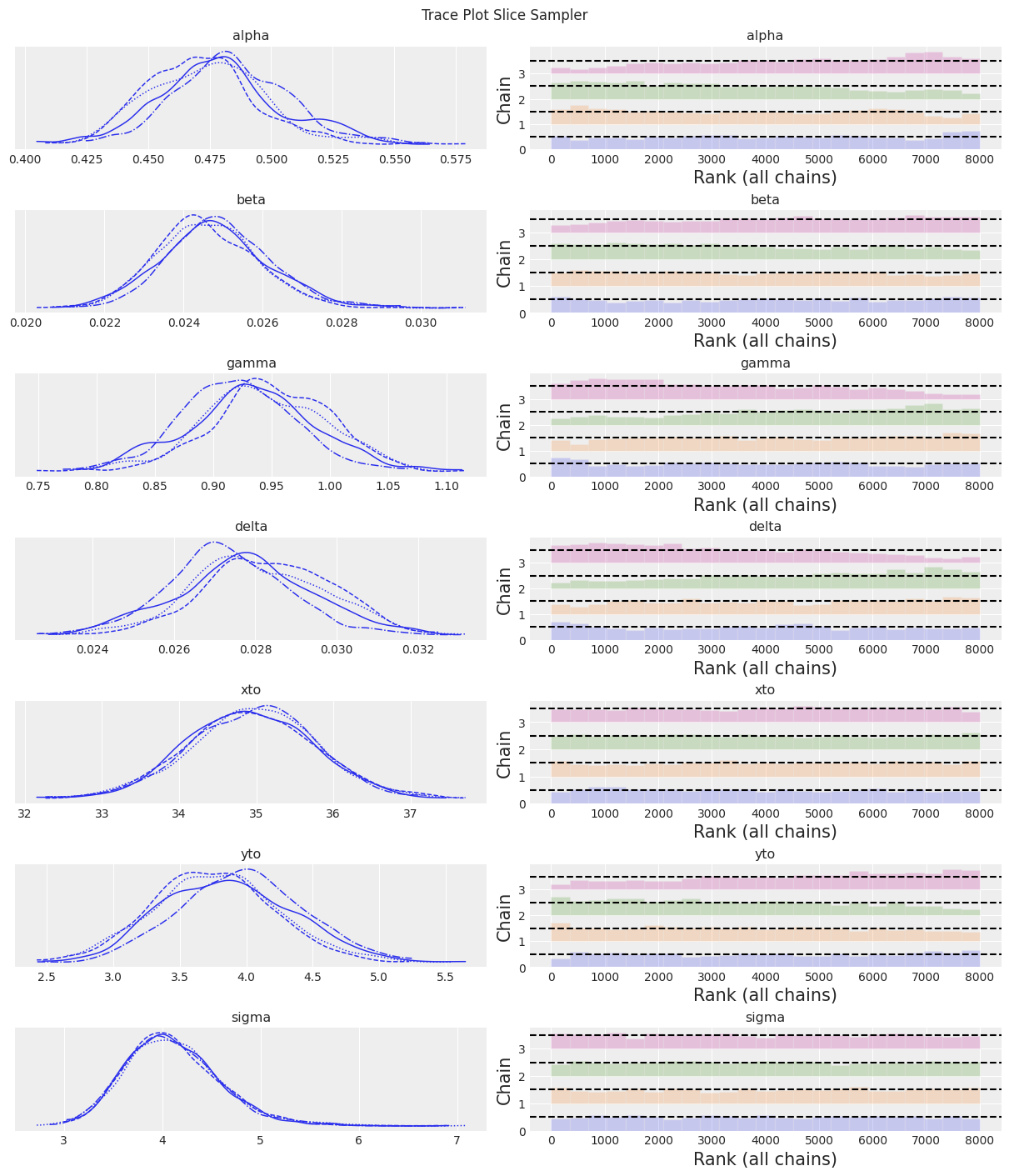
az.plot_trace(trace, kind="rank_bars")
plt.suptitle(f"Trace Plot {sampler}");
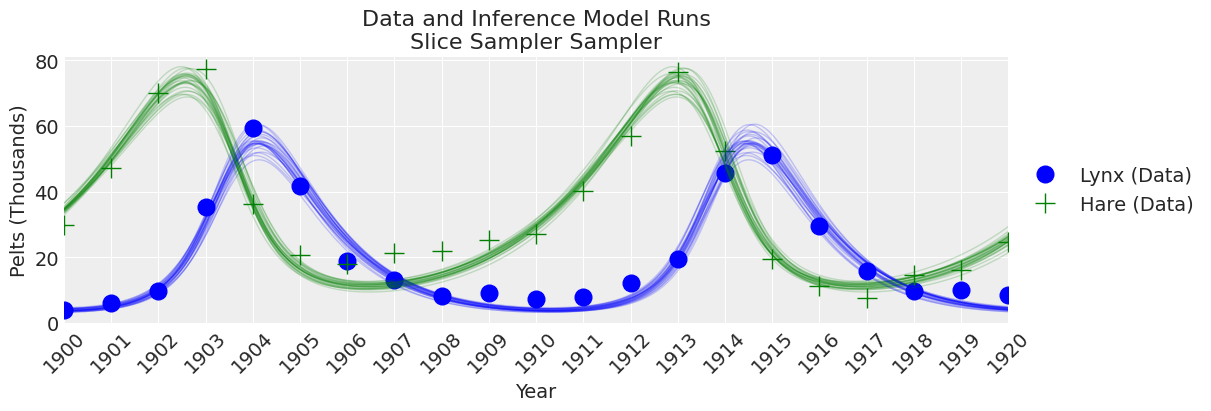
fig, ax = plt.subplots(figsize=(12, 4))
plot_inference(ax, trace, title=f"Data and Inference Model Runs\n{sampler} Sampler");
注释
Slice 采样器速度慢,导致有效样本量较低。 尽管如此,结果开始看起来合理!
DE MetropolisZ 采样器#
sampler = "DEMetropolisZ"
tune = draws = 5000
with model:
trace_DEMZ = pm.sample(step=[pm.DEMetropolisZ(vars_list)], tune=tune, draws=draws)
trace = trace_DEMZ
az.summary(trace)
Multiprocess sampling (4 chains in 4 jobs)
DEMetropolisZ: [alpha, beta, gamma, delta, xto, yto, sigma]
Sampling 4 chains for 5_000 tune and 5_000 draw iterations (20_000 + 20_000 draws total) took 17 seconds.
| 均值 | 标准差 | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 0.482 | 0.024 | 0.434 | 0.523 | 0.001 | 0.001 | 747.0 | 1341.0 | 1.01 |
| beta | 0.025 | 0.001 | 0.022 | 0.028 | 0.000 | 0.000 | 821.0 | 1415.0 | 1.00 |
| gamma | 0.927 | 0.051 | 0.834 | 1.023 | 0.002 | 0.001 | 896.0 | 1547.0 | 1.01 |
| delta | 0.028 | 0.002 | 0.025 | 0.031 | 0.000 | 0.000 | 783.0 | 1432.0 | 1.01 |
| xto | 34.938 | 0.847 | 33.314 | 36.479 | 0.029 | 0.021 | 855.0 | 1201.0 | 1.00 |
| yto | 3.887 | 0.473 | 2.983 | 4.724 | 0.017 | 0.012 | 777.0 | 1156.0 | 1.01 |
| sigma | 4.129 | 0.477 | 3.266 | 5.029 | 0.017 | 0.012 | 799.0 | 1466.0 | 1.00 |
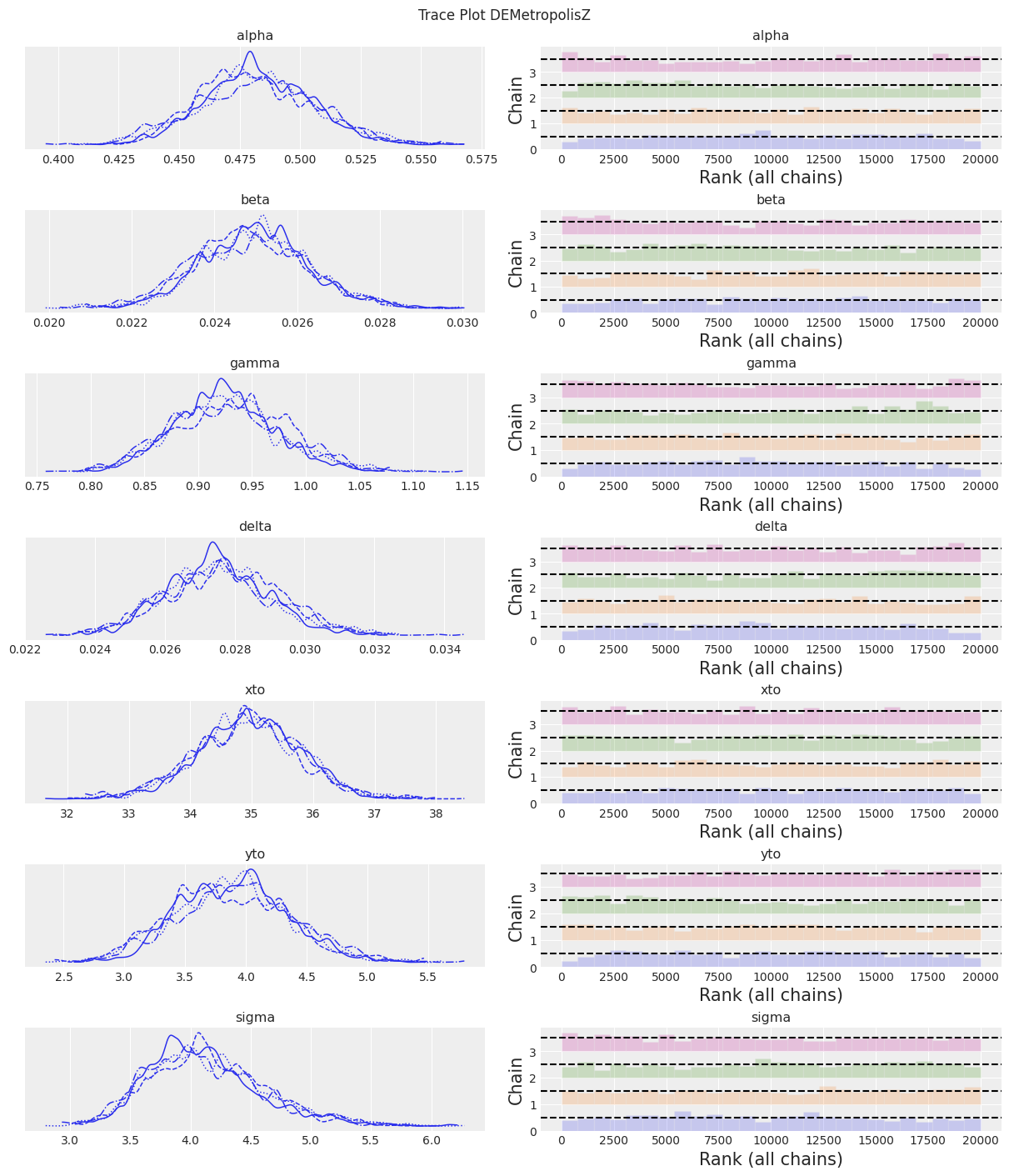
az.plot_trace(trace, kind="rank_bars")
plt.suptitle(f"Trace Plot {sampler}");
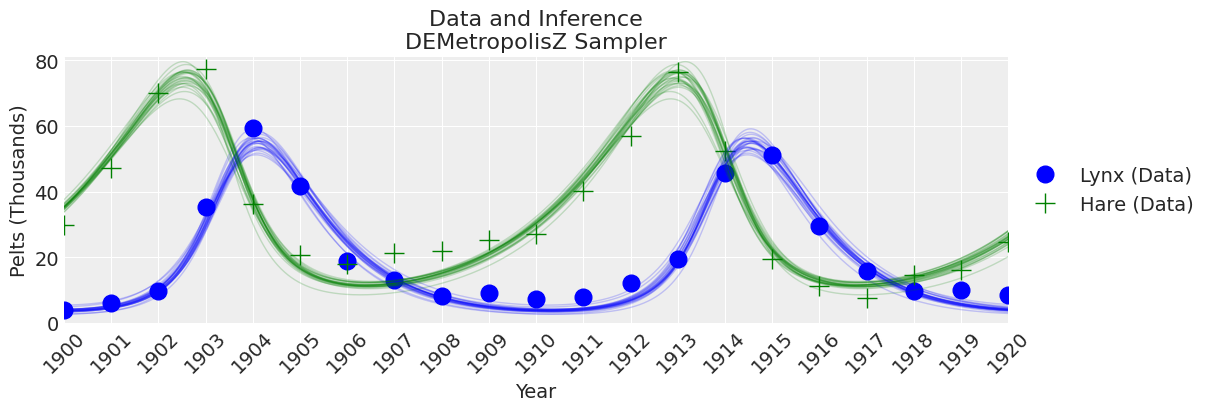
fig, ax = plt.subplots(figsize=(12, 4))
plot_inference(ax, trace, title=f"Data and Inference\n{sampler} Sampler")
注释
DEMetropolisZ 的采样速度比 Slice 采样器快得多,因此每分钟采样的 ESS 更高。 参数估计值相似。 “最终”推断仍然需要增加样本数量。
DEMetropolis 采样器#
在这些实验中,DEMetropolis 采样器不接受 tune,并且要求 chains 至少为 8。 我们将抽取次数设置为 5000,较低的数字(如 3000)会产生不良的混合。
sampler = "DEMetropolis"
chains = 8
draws = 6000
with model:
trace_DEM = pm.sample(step=[pm.DEMetropolis(vars_list)], draws=draws, chains=chains)
trace = trace_DEM
az.summary(trace)
Population sampling (8 chains)
DEMetropolis: [alpha, beta, gamma, delta, xto, yto, sigma]
Attempting to parallelize chains to all cores. You can turn this off with `pm.sample(cores=1)`.
Sampling 8 chains for 1_000 tune and 6_000 draw iterations (8_000 + 48_000 draws total) took 40 seconds.
| 均值 | 标准差 | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 0.483 | 0.021 | 0.443 | 0.520 | 0.000 | 0.000 | 1820.0 | 2647.0 | 1.00 |
| beta | 0.025 | 0.001 | 0.023 | 0.027 | 0.000 | 0.000 | 1891.0 | 3225.0 | 1.00 |
| gamma | 0.924 | 0.045 | 0.837 | 1.008 | 0.001 | 0.001 | 1818.0 | 2877.0 | 1.00 |
| delta | 0.027 | 0.001 | 0.025 | 0.030 | 0.000 | 0.000 | 1628.0 | 2469.0 | 1.00 |
| xto | 34.890 | 0.707 | 33.523 | 36.176 | 0.018 | 0.013 | 1484.0 | 2862.0 | 1.01 |
| yto | 3.897 | 0.403 | 3.126 | 4.644 | 0.010 | 0.007 | 1756.0 | 2468.0 | 1.00 |
| sigma | 4.042 | 0.405 | 3.335 | 4.836 | 0.011 | 0.008 | 1437.0 | 2902.0 | 1.00 |
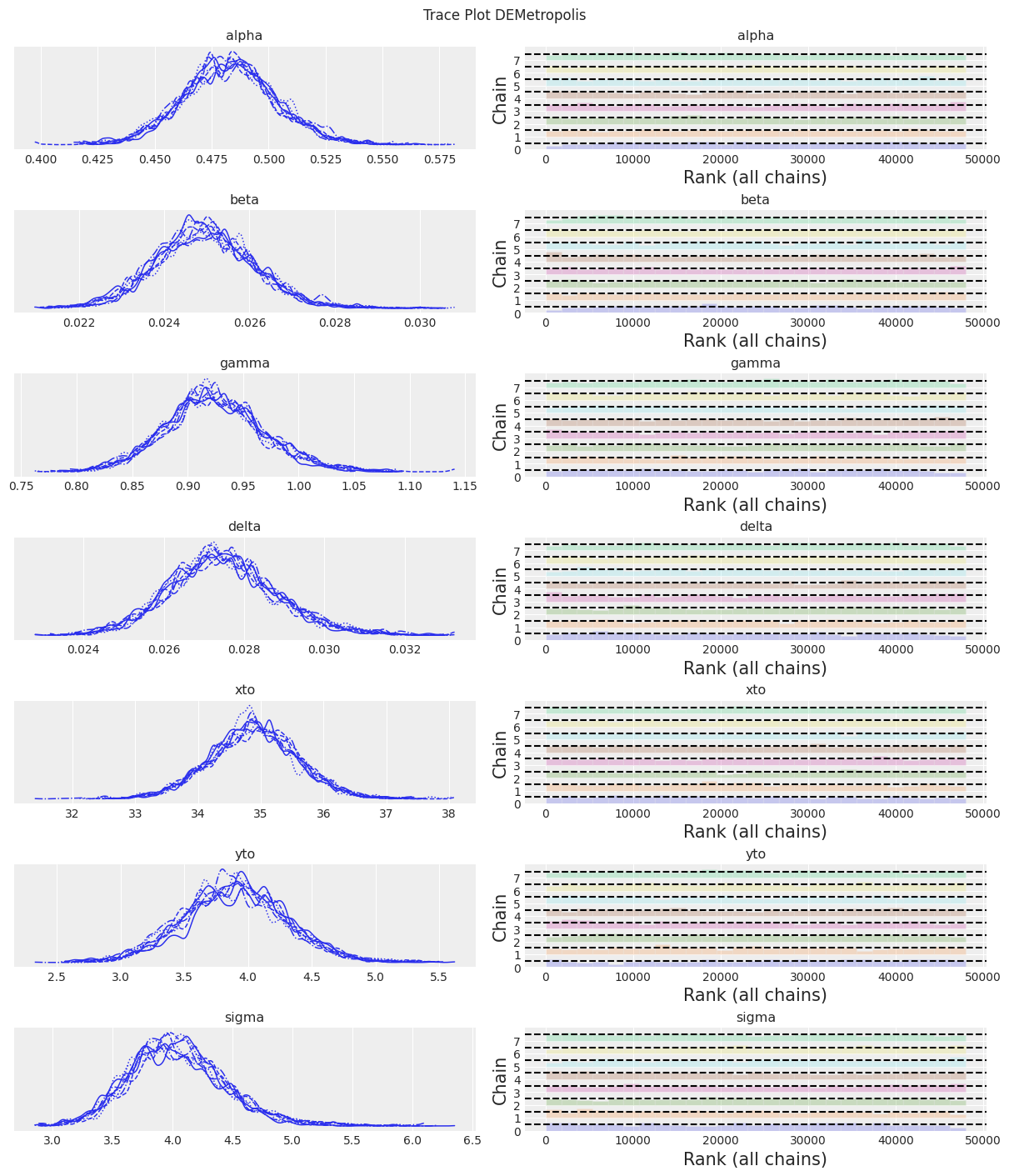
az.plot_trace(trace, kind="rank_bars")
plt.suptitle(f"Trace Plot {sampler}");
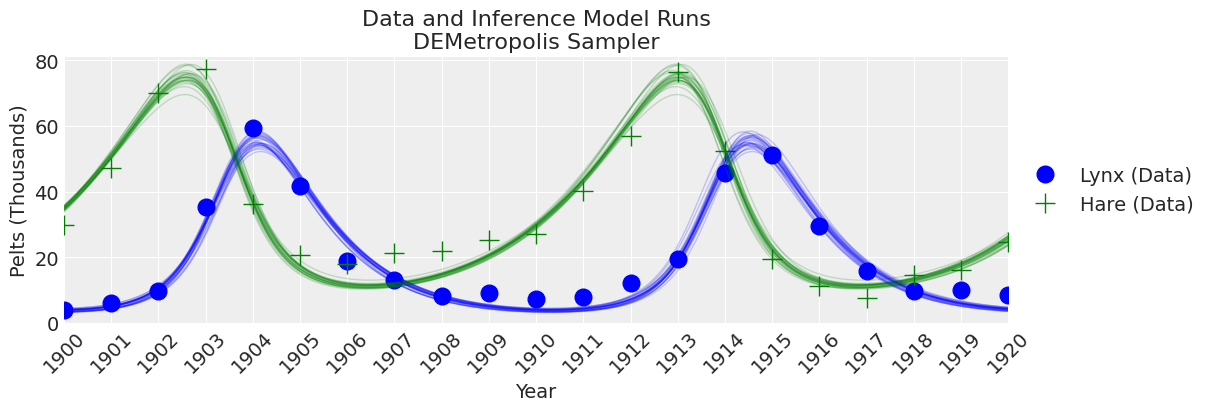
fig, ax = plt.subplots(figsize=(12, 4))
plot_inference(ax, trace, title=f"Data and Inference Model Runs\n{sampler} Sampler");
注释
KDE 看起来太弯曲,但 ESS 很高,R-hat 很好,rank_plots 也看起来不错
Metropolis 采样器#
sampler = "Metropolis"
tune = draws = 5000
with model:
trace_M = pm.sample(step=[pm.Metropolis(vars_list)], tune=tune, draws=draws)
trace = trace_M
az.summary(trace)
Multiprocess sampling (4 chains in 4 jobs)
CompoundStep
>Metropolis: [alpha]
>Metropolis: [beta]
>Metropolis: [gamma]
>Metropolis: [delta]
>Metropolis: [xto]
>Metropolis: [yto]
>Metropolis: [sigma]
/home/osvaldo/anaconda3/envs/pymc/lib/python3.10/site-packages/scipy/integrate/_odepack_py.py:248: ODEintWarning: Excess work done on this call (perhaps wrong Dfun type). Run with full_output = 1 to get quantitative information.
warnings.warn(warning_msg, ODEintWarning)
/home/osvaldo/anaconda3/envs/pymc/lib/python3.10/site-packages/scipy/integrate/_odepack_py.py:248: ODEintWarning: Excess work done on this call (perhaps wrong Dfun type). Run with full_output = 1 to get quantitative information.
warnings.warn(warning_msg, ODEintWarning)
Sampling 4 chains for 5_000 tune and 5_000 draw iterations (20_000 + 20_000 draws total) took 106 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for details
| 均值 | 标准差 | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 0.481 | 0.024 | 0.437 | 0.523 | 0.004 | 0.003 | 44.0 | 112.0 | 1.10 |
| beta | 0.025 | 0.001 | 0.023 | 0.027 | 0.000 | 0.000 | 123.0 | 569.0 | 1.05 |
| gamma | 0.928 | 0.052 | 0.836 | 1.022 | 0.008 | 0.005 | 44.0 | 93.0 | 1.10 |
| delta | 0.028 | 0.002 | 0.025 | 0.031 | 0.000 | 0.000 | 47.0 | 113.0 | 1.09 |
| xto | 34.928 | 0.833 | 33.396 | 36.513 | 0.029 | 0.021 | 808.0 | 1128.0 | 1.00 |
| yto | 3.892 | 0.492 | 3.026 | 4.878 | 0.055 | 0.039 | 81.0 | 307.0 | 1.04 |
| sigma | 4.116 | 0.496 | 3.272 | 5.076 | 0.009 | 0.007 | 2870.0 | 3372.0 | 1.00 |
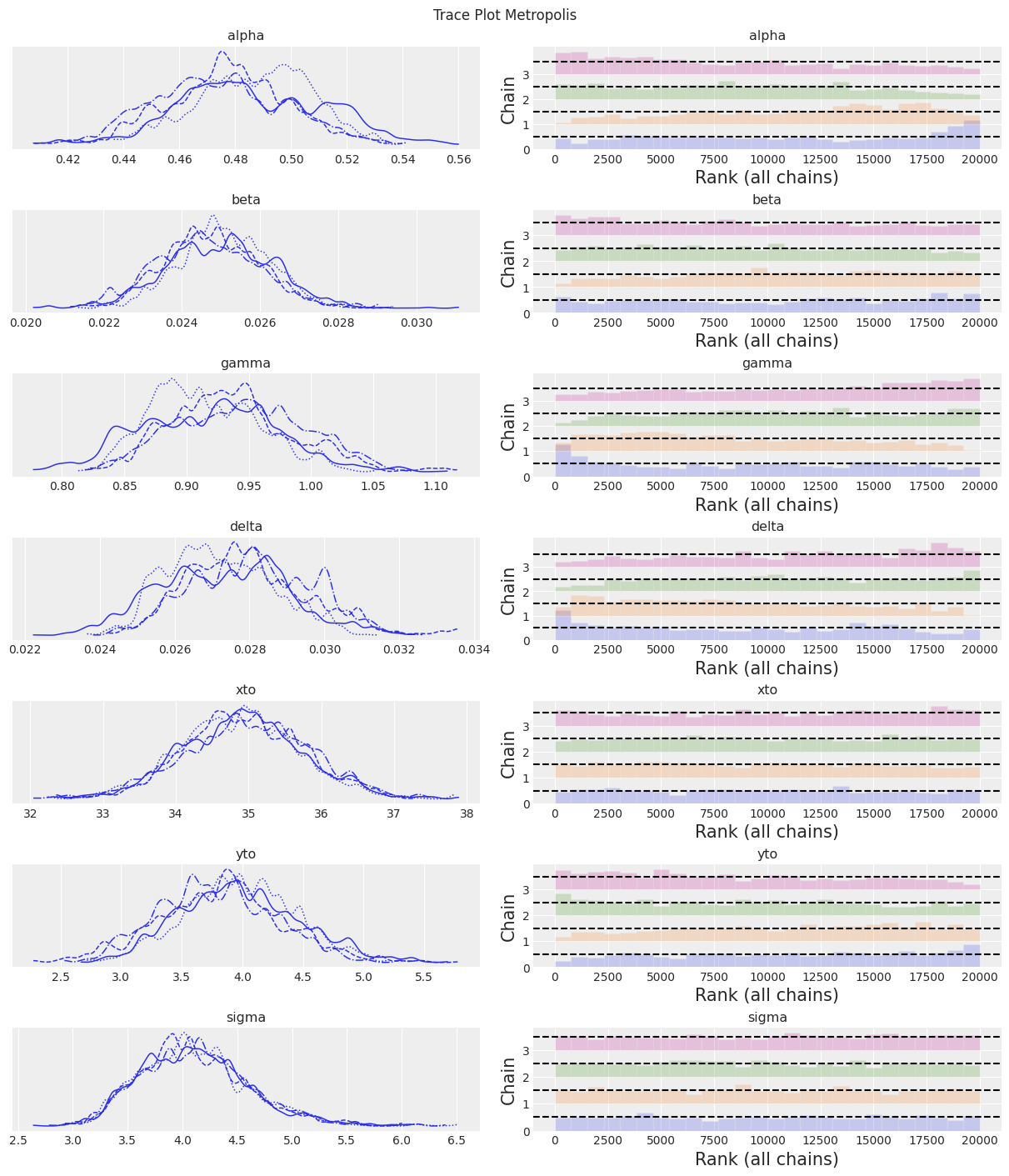
az.plot_trace(trace, kind="rank_bars")
plt.suptitle(f"Trace Plot {sampler}");
fig, ax = plt.subplots(figsize=(12, 4))
plot_inference(ax, trace, title=f"Data and Inference Model Runs\n{sampler} Sampler");
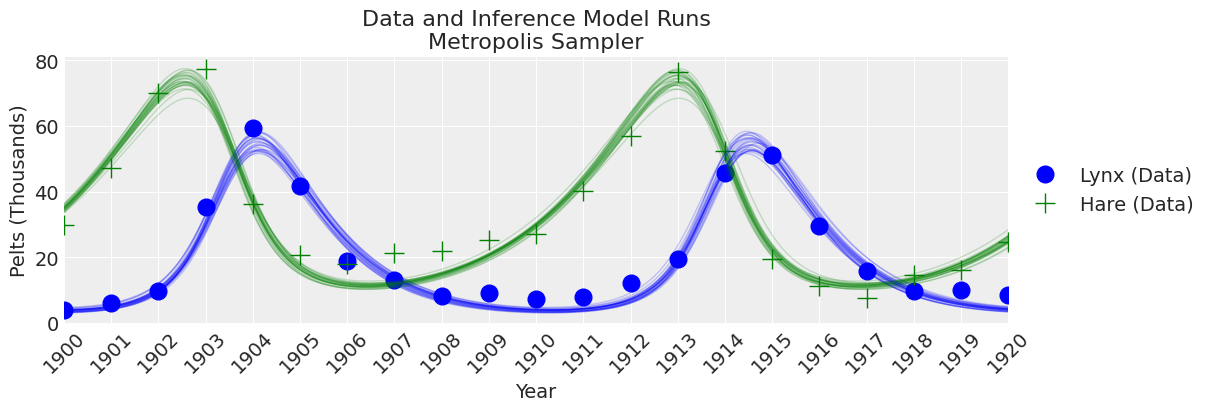
注释
老式的 Metropolis 采样器不如 DEMetroplis 采样器可靠且速度慢。 不推荐使用。
SMC 采样器#
顺序蒙特卡洛 (SMC) 采样器可用于对常规贝叶斯模型进行采样,或在没有似然函数的情况下运行模型(近似贝叶斯计算)。 让我们首先尝试使用常规模型,
具有似然函数的 SMC#
sampler = "SMC with Likelihood"
draws = 2000
with model:
trace_SMC_like = pm.sample_smc(draws)
trace = trace_SMC_like
az.summary(trace)
Initializing SMC sampler...
Sampling 4 chains in 4 jobs
| 均值 | 标准差 | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 0.482 | 0.025 | 0.436 | 0.527 | 0.000 | 0.000 | 8093.0 | 7636.0 | 1.0 |
| beta | 0.025 | 0.001 | 0.022 | 0.027 | 0.000 | 0.000 | 8090.0 | 7582.0 | 1.0 |
| gamma | 0.927 | 0.053 | 0.826 | 1.023 | 0.001 | 0.000 | 8064.0 | 8142.0 | 1.0 |
| delta | 0.028 | 0.002 | 0.025 | 0.031 | 0.000 | 0.000 | 8028.0 | 8016.0 | 1.0 |
| xto | 34.893 | 0.843 | 33.324 | 36.500 | 0.009 | 0.007 | 8060.0 | 7716.0 | 1.0 |
| yto | 3.889 | 0.480 | 2.997 | 4.796 | 0.005 | 0.004 | 7773.0 | 7884.0 | 1.0 |
| sigma | 4.123 | 0.497 | 3.243 | 5.057 | 0.006 | 0.004 | 8169.0 | 7971.0 | 1.0 |
trace.sample_stats._t_sampling
64.09551501274109
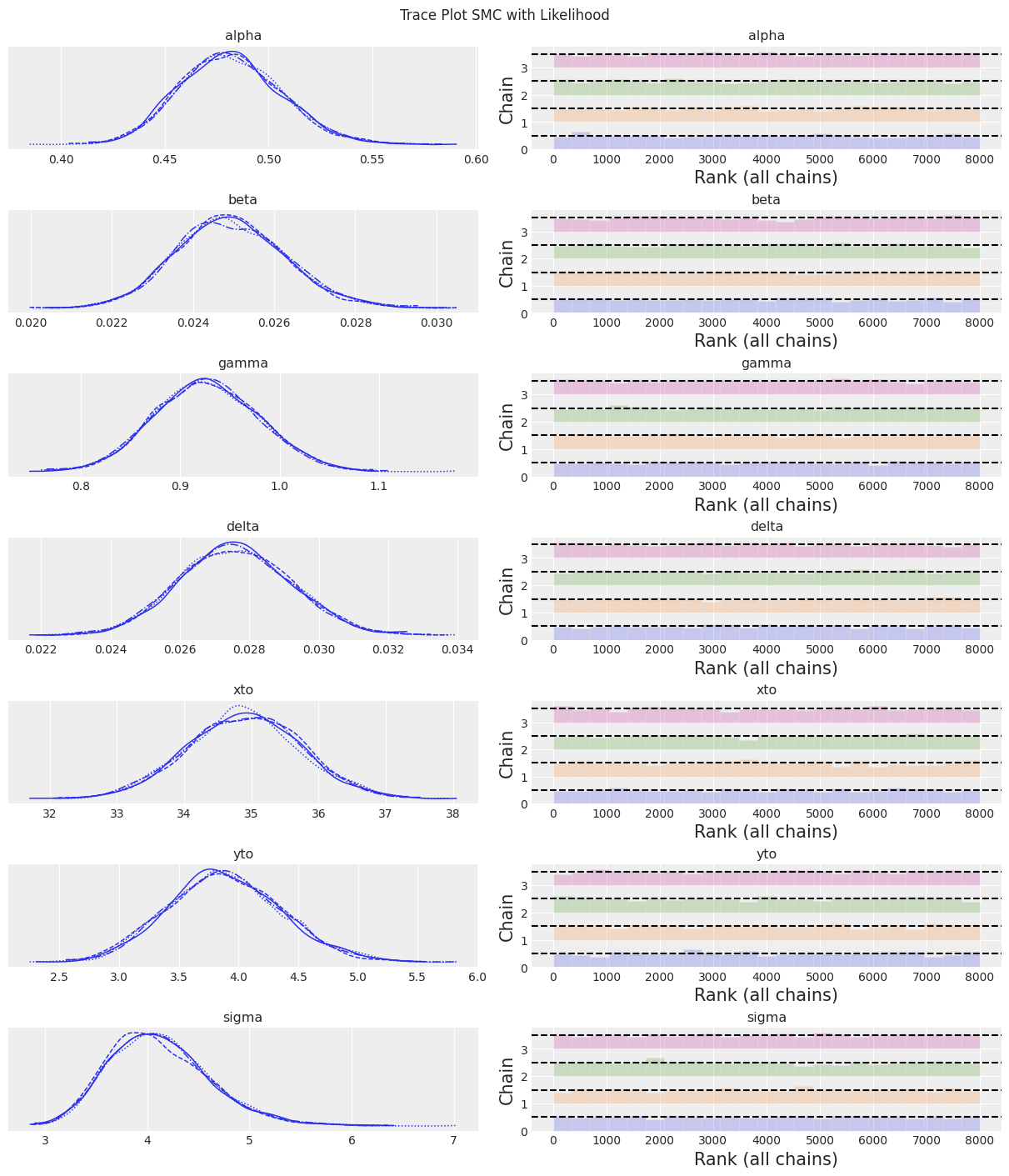
az.plot_trace(trace, kind="rank_bars")
plt.suptitle(f"Trace Plot {sampler}");
fig, ax = plt.subplots(figsize=(12, 4))
plot_inference(ax, trace, title=f"Data and Inference Model Runs\n{sampler} Sampler");
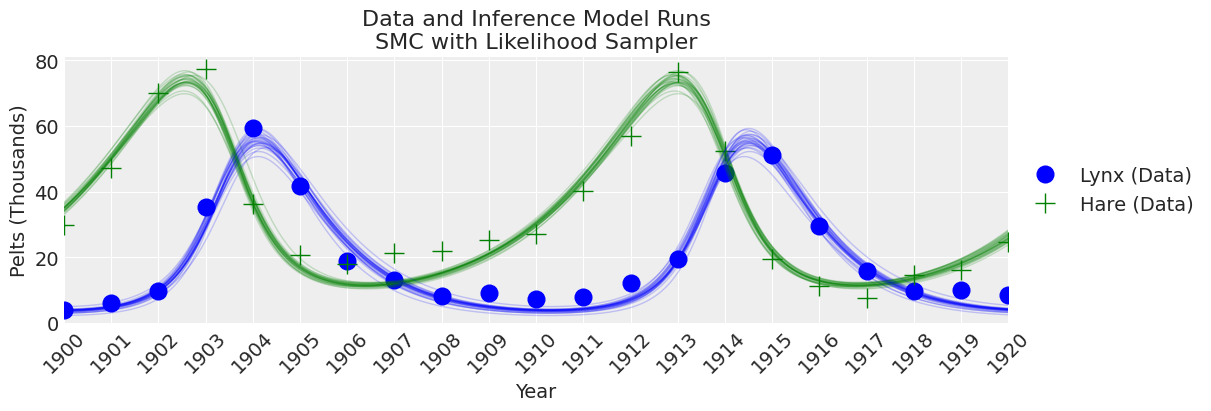
注释
在这种样本数量和调整方案下,与其他采样器相比,SMC 算法导致更宽的不确定性范围。
使用 pm.Simulator Epsilon=1 的 SMC#
如 PyMC.io 上的 SMC 教程中所述,SMC 采样器可用于近似贝叶斯计算,即我们可以使用 pm.Simulator 而不是显式似然函数。 这是用于 SMC-ABC 的 PyMC - odeint 模型的重写。
模拟器函数需要具有正确的签名(例如,首先接受 rng 参数)。
# simulator function based on the signature rng, parameters, size.
def simulator_forward_model(rng, alpha, beta, gamma, delta, xt0, yt0, sigma, size=None):
theta = alpha, beta, gamma, delta, xt0, yt0
mu = odeint(func=rhs, y0=theta[-2:], t=data.year, args=(theta,))
return rng.normal(mu, sigma)
这是带有模拟器函数的模型。 模拟器不是显式似然函数,而是使用模拟值和观测值之间的距离度量(默认为 gaussian)。 使用模拟器时,我们还需要指定 epsilon,即模拟值和观测值之间差异的容差值。 如果 epsilon 太低,SMC 将无法远离初始值或少数几个值。 我们可以通过 az.plot_trace 轻松看到这一点。 如果 epsilon 太高,则后验实际上将是先验。 所以
with pm.Model() as model:
# Specify prior distributions for model parameters
alpha = pm.TruncatedNormal("alpha", mu=theta[0], sigma=0.1, lower=0, initval=theta[0])
beta = pm.TruncatedNormal("beta", mu=theta[1], sigma=0.01, lower=0, initval=theta[1])
gamma = pm.TruncatedNormal("gamma", mu=theta[2], sigma=0.1, lower=0, initval=theta[2])
delta = pm.TruncatedNormal("delta", mu=theta[3], sigma=0.01, lower=0, initval=theta[3])
xt0 = pm.TruncatedNormal("xto", mu=theta[4], sigma=1, lower=0, initval=theta[4])
yt0 = pm.TruncatedNormal("yto", mu=theta[5], sigma=1, lower=0, initval=theta[5])
sigma = pm.HalfNormal("sigma", 10)
# ode_solution
pm.Simulator(
"Y_obs",
simulator_forward_model,
params=(alpha, beta, gamma, delta, xt0, yt0, sigma),
epsilon=1,
observed=data[["hare", "lynx"]].values,
)
推断。 请注意,progressbar 抛出错误,因此已关闭。
sampler = "SMC_epsilon=1"
draws = 2000
with model:
trace_SMC_e1 = pm.sample_smc(draws=draws, progressbar=False)
trace = trace_SMC_e1
az.summary(trace)
Initializing SMC sampler...
Sampling 4 chains in 4 jobs
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for details
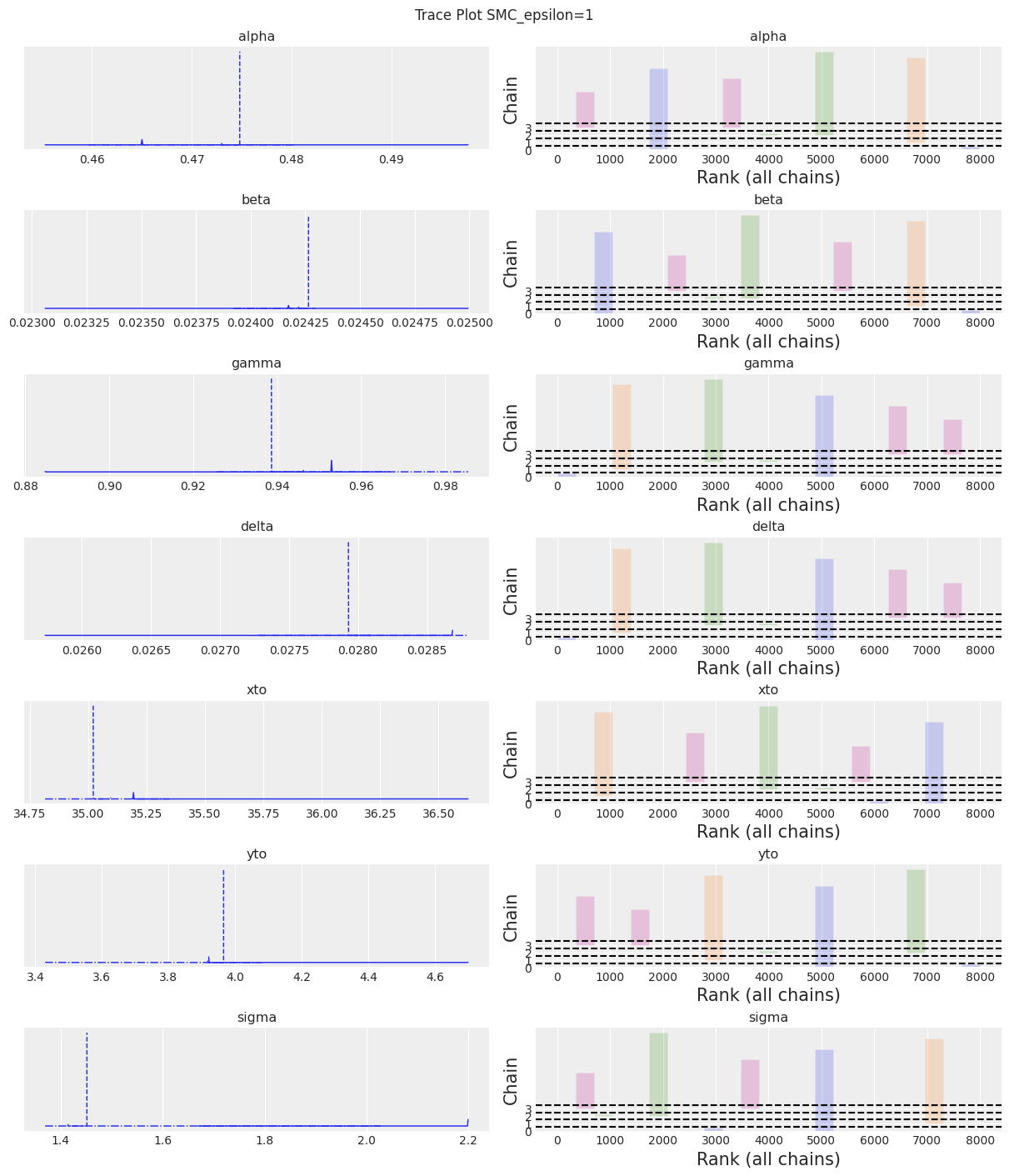
| 均值 | 标准差 | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 0.474 | 0.012 | 0.460 | 0.492 | 0.006 | 0.004 | 5.0 | 5.0 | 3.41 |
| beta | 0.024 | 0.000 | 0.024 | 0.025 | 0.000 | 0.000 | 5.0 | 4.0 | 4.01 |
| gamma | 0.946 | 0.023 | 0.918 | 0.986 | 0.011 | 0.008 | 4.0 | 4.0 | 3.43 |
| delta | 0.028 | 0.001 | 0.028 | 0.029 | 0.000 | 0.000 | 4.0 | 4.0 | 4.19 |
| xto | 34.734 | 0.582 | 33.747 | 35.194 | 0.289 | 0.221 | 4.0 | 4.0 | 7.21 |
| yto | 3.814 | 0.214 | 3.429 | 3.966 | 0.101 | 0.077 | 4.0 | 5.0 | 3.93 |
| sigma | 1.899 | 0.357 | 1.369 | 2.206 | 0.173 | 0.132 | 4.0 | 8000.0 | 4.65 |
az.plot_trace(trace, kind="rank_bars")
plt.suptitle(f"Trace Plot {sampler}");
/home/osvaldo/proyectos/00_BM/arviz/arviz/stats/density_utils.py:487: UserWarning: Your data appears to have a single value or no finite values
warnings.warn("Your data appears to have a single value or no finite values")
fig, ax = plt.subplots(figsize=(12, 4))
plot_inference(ax, trace, title=f"Data and Inference Model Runs\n{sampler} Sampler");
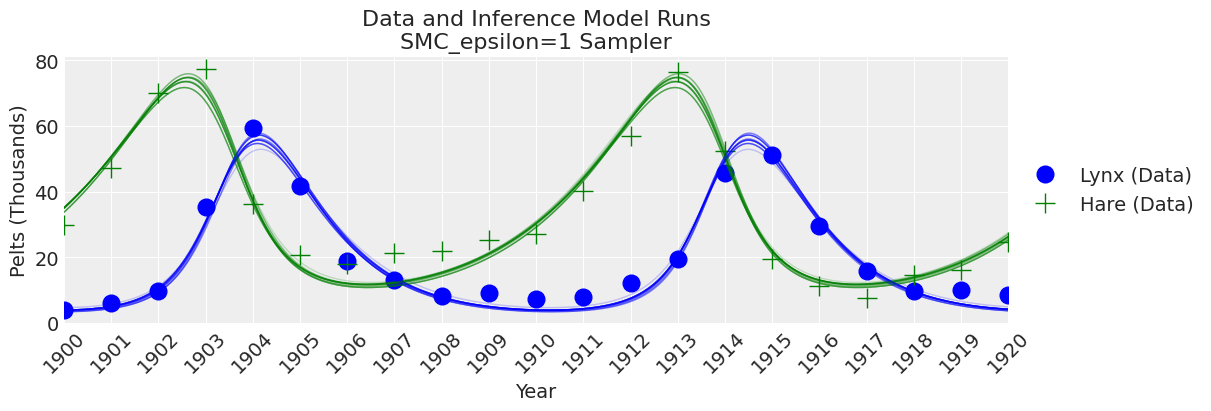
注释
我们可以看到,如果 epsilon 太低,plot_trace 将清楚地显示出来。
epsilon = 10 的 SMC#
with pm.Model() as model:
# Specify prior distributions for model parameters
alpha = pm.TruncatedNormal("alpha", mu=theta[0], sigma=0.1, lower=0, initval=theta[0])
beta = pm.TruncatedNormal("beta", mu=theta[1], sigma=0.01, lower=0, initval=theta[1])
gamma = pm.TruncatedNormal("gamma", mu=theta[2], sigma=0.1, lower=0, initval=theta[2])
delta = pm.TruncatedNormal("delta", mu=theta[3], sigma=0.01, lower=0, initval=theta[3])
xt0 = pm.TruncatedNormal("xto", mu=theta[4], sigma=1, lower=0, initval=theta[4])
yt0 = pm.TruncatedNormal("yto", mu=theta[5], sigma=1, lower=0, initval=theta[5])
sigma = pm.HalfNormal("sigma", 10)
# ode_solution
pm.Simulator(
"Y_obs",
simulator_forward_model,
params=(alpha, beta, gamma, delta, xt0, yt0, sigma),
epsilon=10,
observed=data[["hare", "lynx"]].values,
)
sampler = "SMC epsilon=10"
draws = 2000
with model:
trace_SMC_e10 = pm.sample_smc(draws=draws)
trace = trace_SMC_e10
az.summary(trace)
Initializing SMC sampler...
Sampling 4 chains in 4 jobs
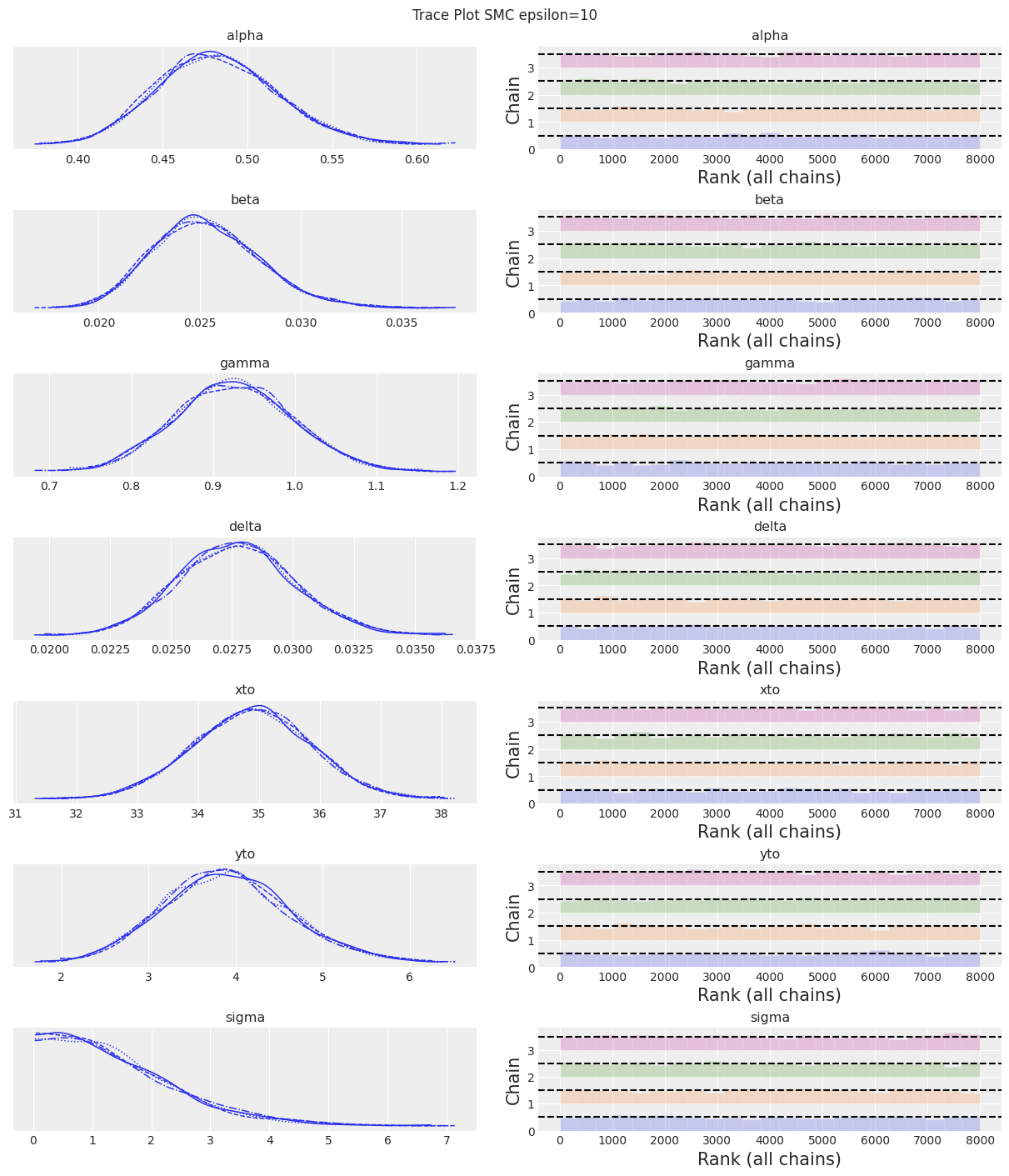
| 均值 | 标准差 | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 0.483 | 0.035 | 0.416 | 0.548 | 0.000 | 0.000 | 7612.0 | 7414.0 | 1.0 |
| beta | 0.025 | 0.003 | 0.020 | 0.030 | 0.000 | 0.000 | 7222.0 | 7768.0 | 1.0 |
| gamma | 0.927 | 0.072 | 0.795 | 1.063 | 0.001 | 0.001 | 7710.0 | 7361.0 | 1.0 |
| delta | 0.028 | 0.002 | 0.023 | 0.032 | 0.000 | 0.000 | 7782.0 | 7565.0 | 1.0 |
| xto | 34.888 | 0.965 | 33.145 | 36.781 | 0.011 | 0.008 | 7921.0 | 7521.0 | 1.0 |
| yto | 3.902 | 0.723 | 2.594 | 5.319 | 0.008 | 0.006 | 7993.0 | 7835.0 | 1.0 |
| sigma | 1.450 | 1.080 | 0.024 | 3.409 | 0.013 | 0.009 | 7490.0 | 7172.0 | 1.0 |
az.plot_trace(trace, kind="rank_bars")
plt.suptitle(f"Trace Plot {sampler}");
fig, ax = plt.subplots(figsize=(12, 4))
plot_inference(ax, trace, title=f"Data and Inference Model Runs\n{sampler} Sampler");
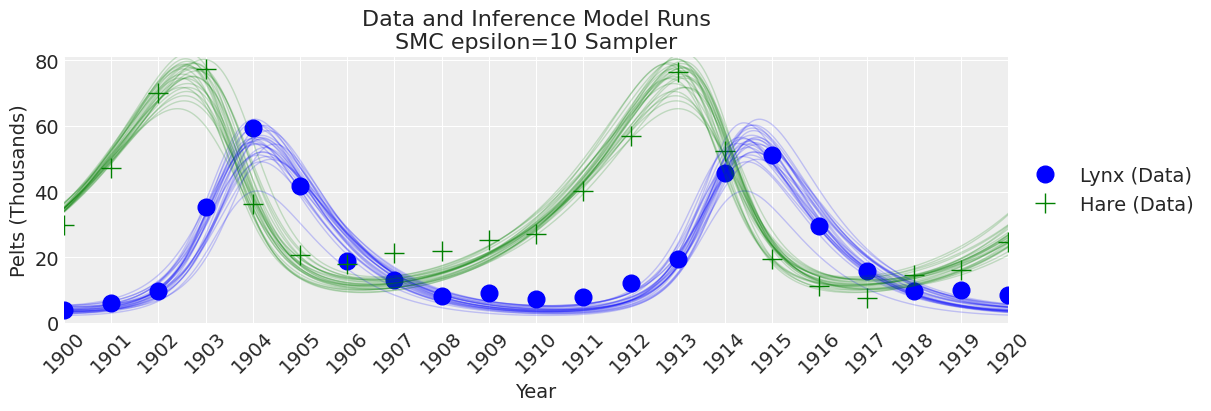
注释
现在我们为 epsilon 设置了更大的值,我们可以看到 SMC 采样器(加上模拟器)提供了良好的结果。 为 epsilon 选择一个值总是会涉及到一些试错。 那么,在实践中该怎么做呢? 由于 epsilon 是距离函数的尺度。 如果您不知道模拟值和观测值之间预期会得到多少误差,那么选择 epsilon 初始猜测的经验法则是使用小于观测数据标准差的数字,可能小一个数量级左右。
后验相关性#
顺便说一句,值得指出的是,后验参数空间对于采样来说是一个困难的几何形状。
az.plot_pair(trace_DEM, figsize=(8, 6), scatter_kwargs=dict(alpha=0.01), marginals=True)
plt.suptitle("Pair Plot Showing Posterior Correlations", size=18);
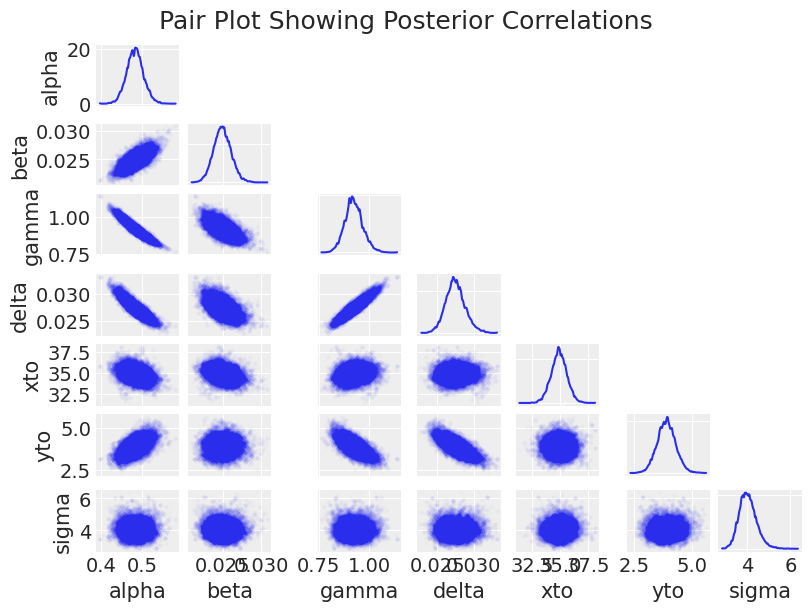
这里的主要观察结果是,对于采样器来说,后验形状非常难以处理,具有正相关、负相关、新月形和尺度上的巨大变化。 这导致了缓慢的采样(除了数千次求解 ODE 的计算开销之外)。 了解模型参数如何相互影响也很有趣。
具有梯度的贝叶斯推断#
只有当向采样器提供梯度时,PyMC 默认采样器 NUTS 才能使用。 在本节中,我们将以两种不同的方式在 PyMC 中求解 ODE 系统,这两种方式都为采样器提供梯度。 第一种是内置的 pymc.ode.DifferentialEquation 求解器,第二种是使用 pytensor.scan 进行前向模拟,这允许循环。 请注意,可能还有其他更好、更快的方法可以使用梯度对 ODE 执行贝叶斯推断,例如 sunode 项目和 diffrax,后者依赖于 JAX。
PyMC ODE 模块#
Pymc.ode 在幕后使用 scipy.odeint 来估计解,然后通过有限差分估计梯度。
pymc.ode API 与 scipy.odeint 类似。 右侧方程放在一个函数中,并编写为 y 和 p 是向量,如下所示。 (即使您的模型有一个状态和/或一个参数,您也应该显式地编写 y[0] 和/或 p[0]。)
def rhs_pymcode(y, t, p):
dX_dt = p[0] * y[0] - p[1] * y[0] * y[1]
dY_dt = -p[2] * y[1] + p[3] * y[0] * y[1]
return [dX_dt, dY_dt]
DifferentialEquation 将以下内容作为参数
func:指定微分方程的函数(即 \(f(\mathbf{y},t,\mathbf{p})\)),times:观察到数据的时刻数组,n_states:\(f(\mathbf{y},t,\mathbf{p})\) 的维度(输出参数的数量),n_theta:\(\mathbf{p}\) 的维度(输入参数的数量),t0:初始条件所属的可选时间,
如下所示
ode_model = DifferentialEquation(
func=rhs_pymcode, times=data.year.values, n_states=2, n_theta=4, t0=data.year.values[0]
)
指定 ODE 后,我们可以在 PyMC 模型中使用它。
使用 NUTS 进行推断#
pymc.ode 速度很慢,因此出于演示目的,我们只会抽取少量样本。
with pm.Model() as model:
# Priors
alpha = pm.TruncatedNormal("alpha", mu=theta[0], sigma=0.1, lower=0, initval=theta[0])
beta = pm.TruncatedNormal("beta", mu=theta[1], sigma=0.01, lower=0, initval=theta[1])
gamma = pm.TruncatedNormal("gamma", mu=theta[2], sigma=0.1, lower=0, initval=theta[2])
delta = pm.TruncatedNormal("delta", mu=theta[3], sigma=0.01, lower=0, initval=theta[3])
xt0 = pm.TruncatedNormal("xto", mu=theta[4], sigma=1, lower=0, initval=theta[4])
yt0 = pm.TruncatedNormal("yto", mu=theta[5], sigma=1, lower=0, initval=theta[5])
sigma = pm.HalfNormal("sigma", 10)
# ode_solution
ode_solution = ode_model(y0=[xt0, yt0], theta=[alpha, beta, gamma, delta])
# Likelihood
pm.Normal("Y_obs", mu=ode_solution, sigma=sigma, observed=data[["hare", "lynx"]].values)
sampler = "NUTS PyMC ODE"
tune = draws = 15
with model:
trace_pymc_ode = pm.sample(tune=tune, draws=draws)
Only 15 samples in chain.
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, beta, gamma, delta, xto, yto, sigma]
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.2340415876362D-14
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.2340415876362D-14
/home/osvaldo/anaconda3/envs/pymc/lib/python3.10/site-packages/scipy/integrate/_odepack_py.py:248: ODEintWarning: Excess work done on this call (perhaps wrong Dfun type). Run with full_output = 1 to get quantitative information.
warnings.warn(warning_msg, ODEintWarning)
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.7324477632756D-17
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.5527744901481D-17
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.1105548980296D-16
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.1105548980296D-16
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.1105548980296D-16
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.1105548980296D-15
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.1105548980296D-15
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.1105548980296D-15
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.1105548980296D-15
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.1105548980296D-14
lsoda-- above warning has been issued i1 times. it will not be issued again for this problem in above message, i1 = 10
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.5241493348134D-15
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.5241493348134D-15
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.1088694571877D-13
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.1088694571877D-13
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.1088694571877D-13
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.4463323525725D-13
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.3388355031231D-13
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.3388355031231D-13
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.3388355031231D-13
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.6776710062462D-13
lsoda-- above warning has been issued i1 times. it will not be issued again for this problem in above message, i1 = 10
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.4158457835953D-42
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.4158457835953D-42
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.1370912617246D-40
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.1370912617246D-40
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.1370912617246D-40
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.5948148309049D-40
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.4374718724784D-40
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.3628059832750D-40
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.3628059832750D-40
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.3628059832750D-40
lsoda-- above warning has been issued i1 times. it will not be issued again for this problem in above message, i1 = 10
/home/osvaldo/anaconda3/envs/pymc/lib/python3.10/site-packages/scipy/integrate/_odepack_py.py:248: ODEintWarning: Excess work done on this call (perhaps wrong Dfun type). Run with full_output = 1 to get quantitative information.
warnings.warn(warning_msg, ODEintWarning)
/home/osvaldo/anaconda3/envs/pymc/lib/python3.10/site-packages/scipy/integrate/_odepack_py.py:248: ODEintWarning: Excess work done on this call (perhaps wrong Dfun type). Run with full_output = 1 to get quantitative information.
warnings.warn(warning_msg, ODEintWarning)
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.8146615298522D-82
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.8146615298522D-82
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.8146615298522D-78
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.8146615298522D-78
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.8146615298522D-78
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.8146615298522D-77
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.8146615298522D-77
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.8146615298522D-77
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.8146615298522D-77
lsoda-- warning..internal t (=r1) and h (=r2) are such that in the machine, t + h = t on the next step
(h = step size). solver will continue anyway in above, r1 = 0.1900000000000D+04 r2 = 0.7775771408140D-76
lsoda-- above warning has been issued i1 times. it will not be issued again for this problem in above message, i1 = 10
Sampling 4 chains for 15 tune and 15 draw iterations (60 + 60 draws total) took 60 seconds.
The number of samples is too small to check convergence reliably.
trace = trace_pymc_ode
az.summary(trace)
| 均值 | 标准差 | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 0.472 | 0.031 | 0.389 | 0.506 | 0.008 | 0.006 | 17.0 | 33.0 | 1.36 |
| beta | 0.026 | 0.003 | 0.022 | 0.032 | 0.001 | 0.001 | 12.0 | 40.0 | 1.54 |
| gamma | 0.959 | 0.080 | 0.868 | 1.151 | 0.025 | 0.018 | 11.0 | 33.0 | 1.59 |
| delta | 0.029 | 0.003 | 0.026 | 0.035 | 0.001 | 0.001 | 14.0 | 37.0 | 1.33 |
| xto | 34.907 | 0.852 | 33.526 | 36.300 | 0.099 | 0.071 | 98.0 | 43.0 | 1.21 |
| yto | 3.347 | 0.772 | 1.742 | 4.342 | 0.278 | 0.205 | 10.0 | 16.0 | 1.78 |
| sigma | 6.117 | 4.425 | 3.502 | 16.420 | 1.353 | 0.984 | 9.0 | 16.0 | 1.87 |
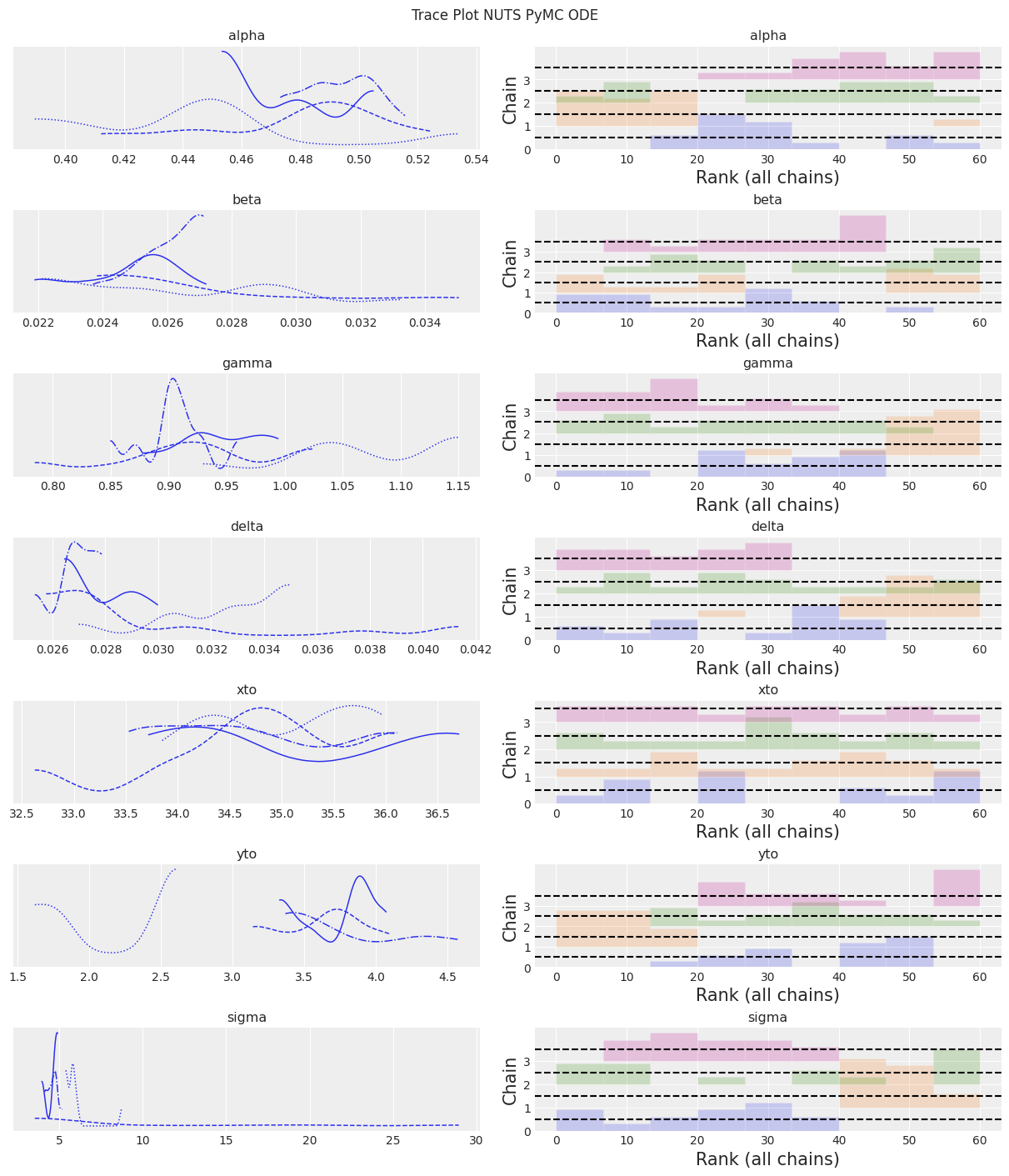
az.plot_trace(trace, kind="rank_bars")
plt.suptitle(f"Trace Plot {sampler}");
_, ax = plt.subplots(figsize=(12, 4))
plot_inference(ax, trace, title=f"Data and Inference Model Runs\n{sampler} Sampler");
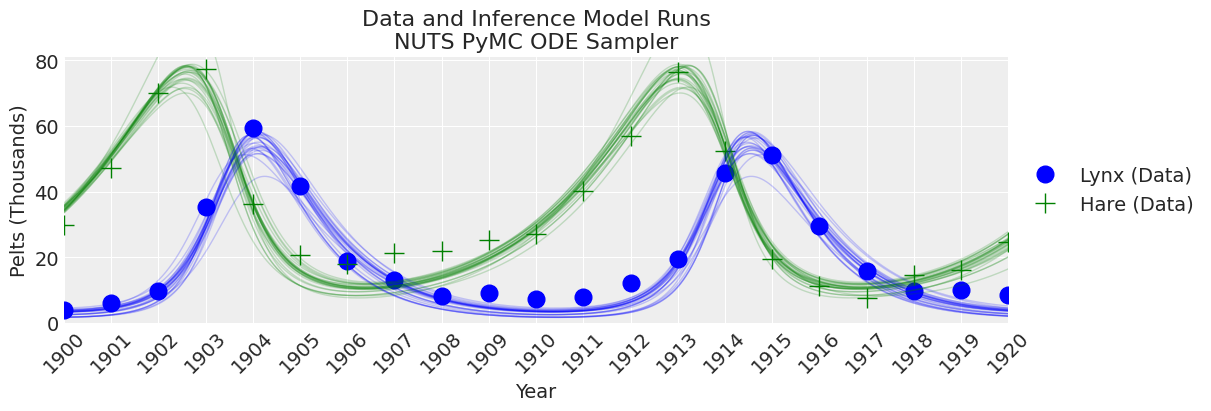
注释
NUTS 开始找到正确的后验,但需要更多时间才能做出良好的推断。
使用 Pytensor Scan 进行模拟#
最后,我们可以将 ODE 系统编写为 PyMC 中的前向模拟求解器。 在 PyMC 中编写 for 循环的方法是使用 pytensor.scan. 然后通过自动微分将梯度提供给采样器。
首先,我们应该测试时间步长是否足够小以获得合理的估计。
检查时间步长#
创建一个接受不同时间步长数量的函数以进行测试。 该函数还演示了如何使用 pytensor.scan。
# Lotka-Volterra forward simulation model using scan
def lv_scan_simulation_model(theta, steps_year=100, years=21):
# variables to control time steps
n_steps = years * steps_year
dt = 1 / steps_year
# PyMC model
with pm.Model() as model:
# Priors (these are static for testing)
alpha = theta[0]
beta = theta[1]
gamma = theta[2]
delta = theta[3]
xt0 = theta[4]
yt0 = theta[5]
# Lotka-Volterra calculation function
## Similar to the right-hand-side functions used earlier
## but with dt applied to the equations
def ode_update_function(x, y, alpha, beta, gamma, delta):
x_new = x + (alpha * x - beta * x * y) * dt
y_new = y + (-gamma * y + delta * x * y) * dt
return x_new, y_new
# Pytensor scan looping function
## The function argument names are not intuitive in this context!
result, updates = pytensor.scan(
fn=ode_update_function, # function
outputs_info=[xt0, yt0], # initial conditions
non_sequences=[alpha, beta, gamma, delta], # parameters
n_steps=n_steps, # number of loops
)
# Put the results together and track the result
pm.Deterministic("result", pm.math.stack([result[0], result[1]], axis=1))
return model
运行各种时间步长的模拟并绘制结果。
_, ax = plt.subplots(figsize=(12, 4))
steps_years = [12, 100, 1000, 10000]
for steps_year in steps_years:
time = np.arange(1900, 1921, 1 / steps_year)
model = lv_scan_simulation_model(theta, steps_year=steps_year)
with model:
prior = pm.sample_prior_predictive(1)
ax.plot(time, prior.prior.result[0][0].values, label=str(steps_year) + " steps/year")
ax.legend(loc="center left", bbox_to_anchor=(1, 0.5))
ax.set_title("Lotka-Volterra Forward Simulation Model with different step sizes");
Sampling: []
Sampling: []
Sampling: []
Sampling: []
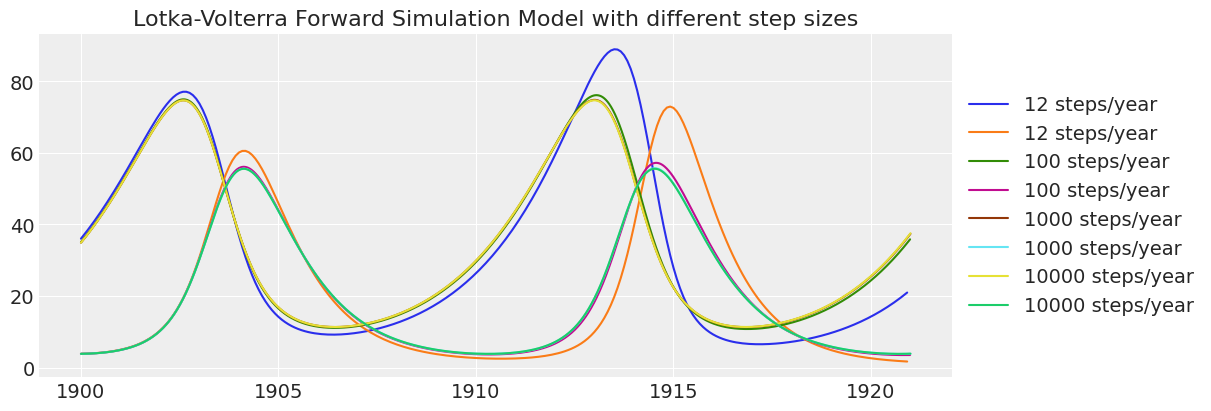
请注意,较低分辨率的模拟随着时间的推移精度会降低。 基于此检查,每年 100 个时间步长就足够准确了。 每年 12 个步长在 20 年的模拟中具有过多的“数值扩散”。
使用 NUTS 进行推断#
现在我们对每年 100 个时间步长感到满意,我们编写带有索引的模型以将数据与结果对齐。
def lv_scan_inference_model(theta, steps_year=100, years=21):
# variables to control time steps
n_steps = years * steps_year
dt = 1 / steps_year
# variables to control indexing to get annual values
segment = [True] + [False] * (steps_year - 1)
boolist_idxs = []
for _ in range(years):
boolist_idxs += segment
# PyMC model
with pm.Model() as model:
# Priors
alpha = pm.TruncatedNormal("alpha", mu=theta[0], sigma=0.1, lower=0, initval=theta[0])
beta = pm.TruncatedNormal("beta", mu=theta[1], sigma=0.01, lower=0, initval=theta[1])
gamma = pm.TruncatedNormal("gamma", mu=theta[2], sigma=0.1, lower=0, initval=theta[2])
delta = pm.TruncatedNormal("delta", mu=theta[3], sigma=0.01, lower=0, initval=theta[3])
xt0 = pm.TruncatedNormal("xto", mu=theta[4], sigma=1, lower=0, initval=theta[4])
yt0 = pm.TruncatedNormal("yto", mu=theta[5], sigma=1, lower=0, initval=theta[5])
sigma = pm.HalfNormal("sigma", 10)
# Lotka-Volterra calculation function
def ode_update_function(x, y, alpha, beta, gamma, delta):
x_new = x + (alpha * x - beta * x * y) * dt
y_new = y + (-gamma * y + delta * x * y) * dt
return x_new, y_new
# Pytensor scan is a looping function
result, updates = pytensor.scan(
fn=ode_update_function, # function
outputs_info=[xt0, yt0], # initial conditions
non_sequences=[alpha, beta, gamma, delta], # parameters
n_steps=n_steps,
) # number of loops
# Put the results together
final_result = pm.math.stack([result[0], result[1]], axis=1)
# Filter the results down to annual values
annual_value = final_result[np.array(boolist_idxs), :]
# Likelihood function
pm.Normal("Y_obs", mu=annual_value, sigma=sigma, observed=data[["hare", "lynx"]].values)
return model
这也非常慢,因此我们只抽取少量样本用于演示目的。
steps_year = 100
model = lv_scan_inference_model(theta, steps_year=steps_year)
sampler = "NUTS Pytensor Scan"
tune = draws = 50
with model:
trace_scan = pm.sample(tune=tune, draws=draws)
Only 50 samples in chain.
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [alpha, beta, gamma, delta, xto, yto, sigma]
Sampling 4 chains for 50 tune and 50 draw iterations (200 + 200 draws total) took 89 seconds.
The number of samples is too small to check convergence reliably.
trace = trace_scan
az.summary(trace)
| 均值 | 标准差 | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 0.480 | 0.025 | 0.432 | 0.526 | 0.003 | 0.002 | 77.0 | 94.0 | 1.02 |
| beta | 0.025 | 0.001 | 0.023 | 0.027 | 0.000 | 0.000 | 147.0 | 155.0 | 1.03 |
| gamma | 0.933 | 0.054 | 0.832 | 1.030 | 0.007 | 0.005 | 70.0 | 80.0 | 1.04 |
| delta | 0.028 | 0.002 | 0.024 | 0.031 | 0.000 | 0.000 | 70.0 | 94.0 | 1.04 |
| xto | 34.877 | 0.764 | 33.232 | 36.118 | 0.046 | 0.032 | 265.0 | 110.0 | 1.04 |
| yto | 3.987 | 0.504 | 2.887 | 4.749 | 0.069 | 0.049 | 58.0 | 102.0 | 1.06 |
| sigma | 4.173 | 0.488 | 3.361 | 5.005 | 0.056 | 0.039 | 83.0 | 104.0 | 1.03 |
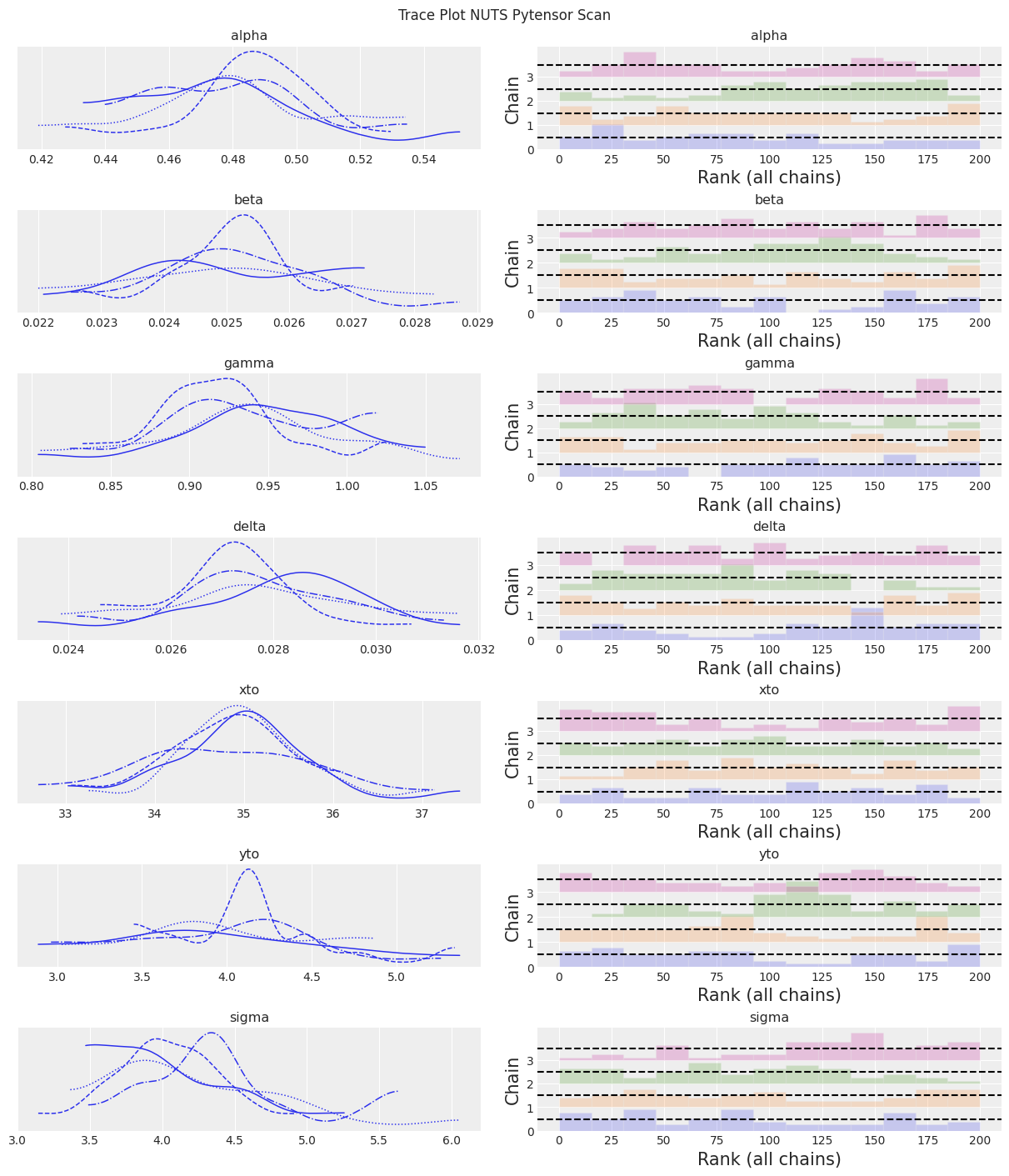
az.plot_trace(trace, kind="rank_bars")
plt.suptitle(f"Trace Plot {sampler}");
(2100, 2)
_, ax = plt.subplots(figsize=(12, 4))
plot_inference(ax, trace, title=f"Data and Inference Model Runs\n{sampler} Sampler");
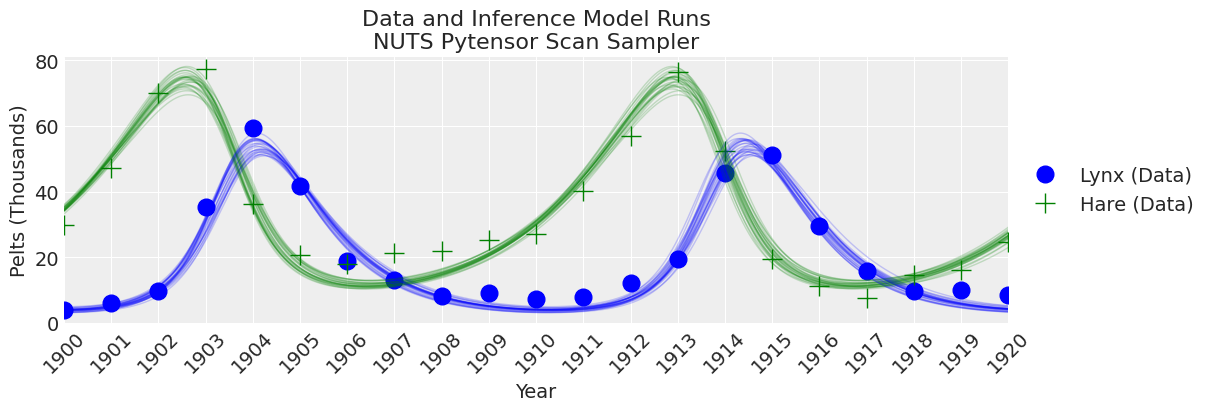
注释
采样器比 pymc.ode 实现更快,但仍然比 scipy odeint 结合无梯度推断方法慢。
总结#
让我们比较这些不同方法之间的推断结果。 回想一下,为了在合理的时间内运行此笔记本,我们对许多推断方法的样本数量不足。 为了进行公平的比较,我们需要增加样本数量并运行笔记本更长时间。 无论如何,让我们看一下。
# Make lists with variable for looping
var_names = [str(s).split("_")[0] for s in list(model.values_to_rvs.keys())[:-1]]
# Make lists with model results and model names for plotting
inference_results = [
trace_slice,
trace_DEMZ,
trace_DEM,
trace_M,
trace_SMC_like,
trace_SMC_e1,
trace_SMC_e10,
trace_pymc_ode,
trace_scan,
]
model_names = [
"Slice Sampler",
"DEMetropolisZ",
"DEMetropolis",
"Metropolis",
"SMC with Likelihood",
"SMC e=1",
"SMC e=10",
"PyMC ODE NUTs",
"Pytensor Scan NUTs",
]
# Loop through variable names
for var_name in var_names:
axes = az.plot_forest(
inference_results,
model_names=model_names,
var_names=var_name,
kind="forestplot",
legend=False,
combined=True,
figsize=(7, 3),
)
axes[0].set_title(f"Marginal Probability: {var_name}")
# Clean up ytick labels
ylabels = axes[0].get_yticklabels()
new_ylabels = []
for label in ylabels:
txt = label.get_text()
txt = txt.replace(": " + var_name, "")
label.set_text(txt)
new_ylabels.append(label)
axes[0].set_yticklabels(new_ylabels)
plt.show();
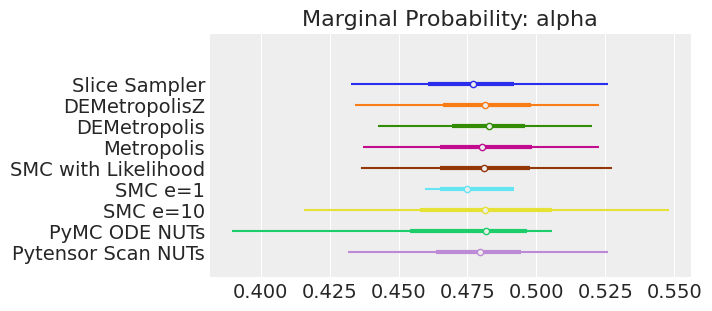
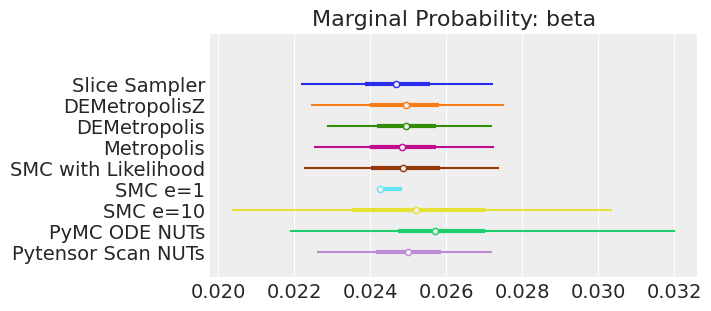
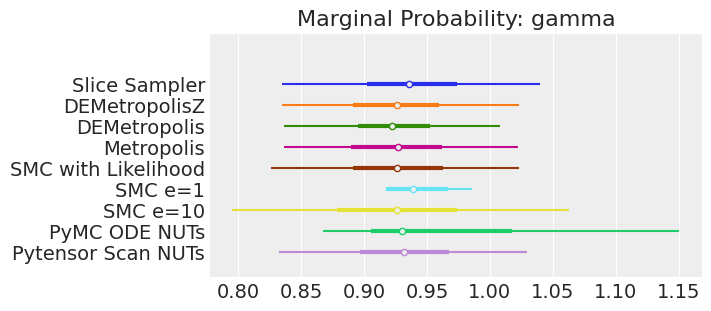
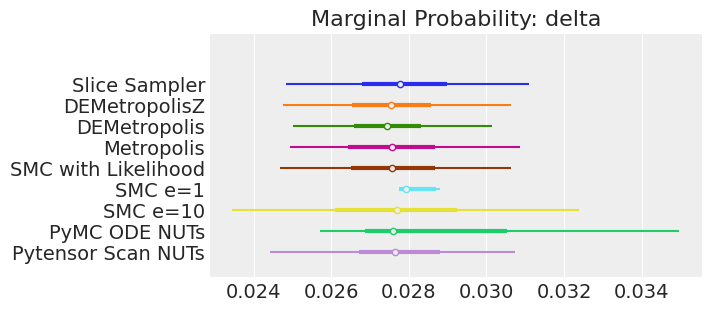
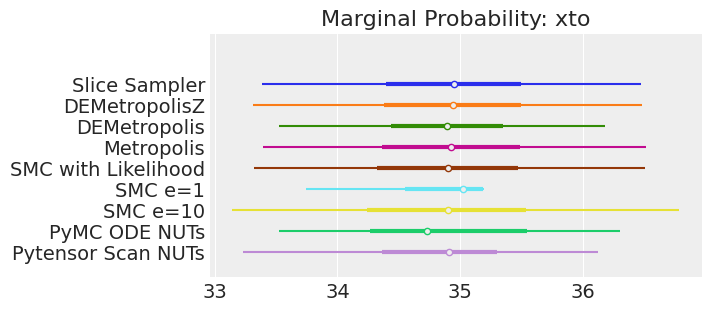
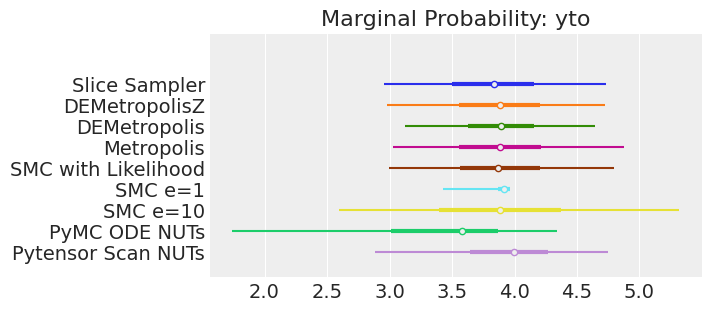
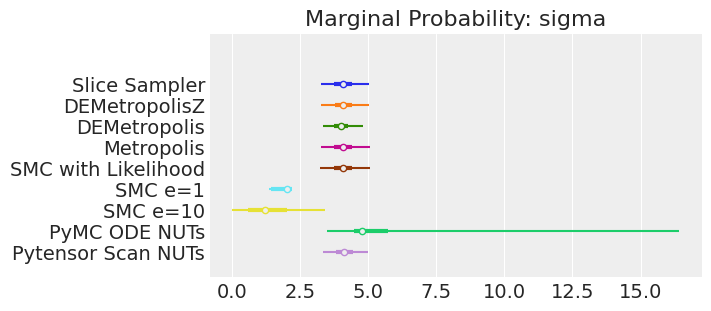
注释
如果我们运行采样器足够长的时间以获得良好的推断,我们预计它们会收敛到相同的后验概率分布。 对于近似贝叶斯计算来说,情况不一定如此,除非我们首先确保对似然的近似足够好。 例如,SMCe=1 提供了错误的结果,当我们使用 plot_trace 作为诊断时,我们一直在警告这很可能是这种情况。 对于 SMC e=10,我们看到后验均值与其他采样器一致,但后验更宽。 这是 ABC 方法的预期结果。 较小的 epsilon 值(可能为 5)应该提供更接近真实值的后验。
主要结论#
我们以 4 种主要方式对 ODE 系统执行了贝叶斯推断
Scipy
odeint包装在 Pytensorop中,并使用无梯度采样器进行采样(比较 5 种不同的采样器)。Scipy
odeint包装在pm.Simulator函数中,并使用非基于似然的顺序蒙特卡洛 (SMC) 采样器进行采样。PyMC
ode.DifferentialEquation使用 NUTS 进行采样。使用
pytensor.scan进行前向模拟,并使用 NUTS 进行采样。
对于这个问题,“赢家”是 Scipy odeint 求解器,它与微分进化 (DE) Metropolis 采样器和 SMC(对于具有似然函数的模型)结合使用,提供了良好的结果,SMC 稍慢一些(但诊断效果也更好)。 NUTS 采样器效率的提高并没有弥补使用带有梯度的慢速 ODE 求解器的效率低下。 DEMetropolis 和 SMC 为具有工作数值模型并渴望执行贝叶斯推断的科学家提供了最简单的工作流程。 只是将数值模型包装在 Pytensor op 中并将其插入 PyMC 模型中,就可以让您走得很远!
参考文献#
Richard McElreath. *Statistical rethinking: A Bayesian course with examples in R and Stan*。 Chapman and Hall/CRC, 2018。
水印#
%watermark -n -u -v -iv -w
Last updated: Thu Mar 30 2023
Python implementation: CPython
Python version : 3.10.10
IPython version : 8.10.0
pytensor : 2.10.1
pandas : 1.5.3
matplotlib: 3.5.2
pymc : 5.1.2+12.g67925df69
numpy : 1.23.5
arviz : 0.16.0.dev0
Watermark: 2.3.1
许可声明#
本示例库中的所有笔记本均根据 MIT 许可证 提供,该许可证允许修改和再分发以用于任何用途,前提是保留版权和许可声明。
引用 PyMC 示例#
要引用此笔记本,请使用 Zenodo 为 pymc-examples 存储库提供的 DOI。
重要提示
许多笔记本改编自其他来源:博客、书籍... 在这种情况下,您也应该引用原始来源。
另请记住引用您的代码使用的相关库。
这是一个 bibtex 中的引用模板
@incollection{citekey,
author = "<notebook authors, see above>",
title = "<notebook title>",
editor = "PyMC Team",
booktitle = "PyMC examples",
doi = "10.5281/zenodo.5654871"
}
一旦渲染,它可能看起来像